An Overview of Recent Developments in the Application of Antigen Displaying Vaccine Platforms: Hints for Future SARS-CoV-2 VLP Vaccines
Abstract
:1. Introduction
2. Various Protein Expression Platforms for VLPs
3. Genetic Display of Subunit Antigens on VLP Carriers
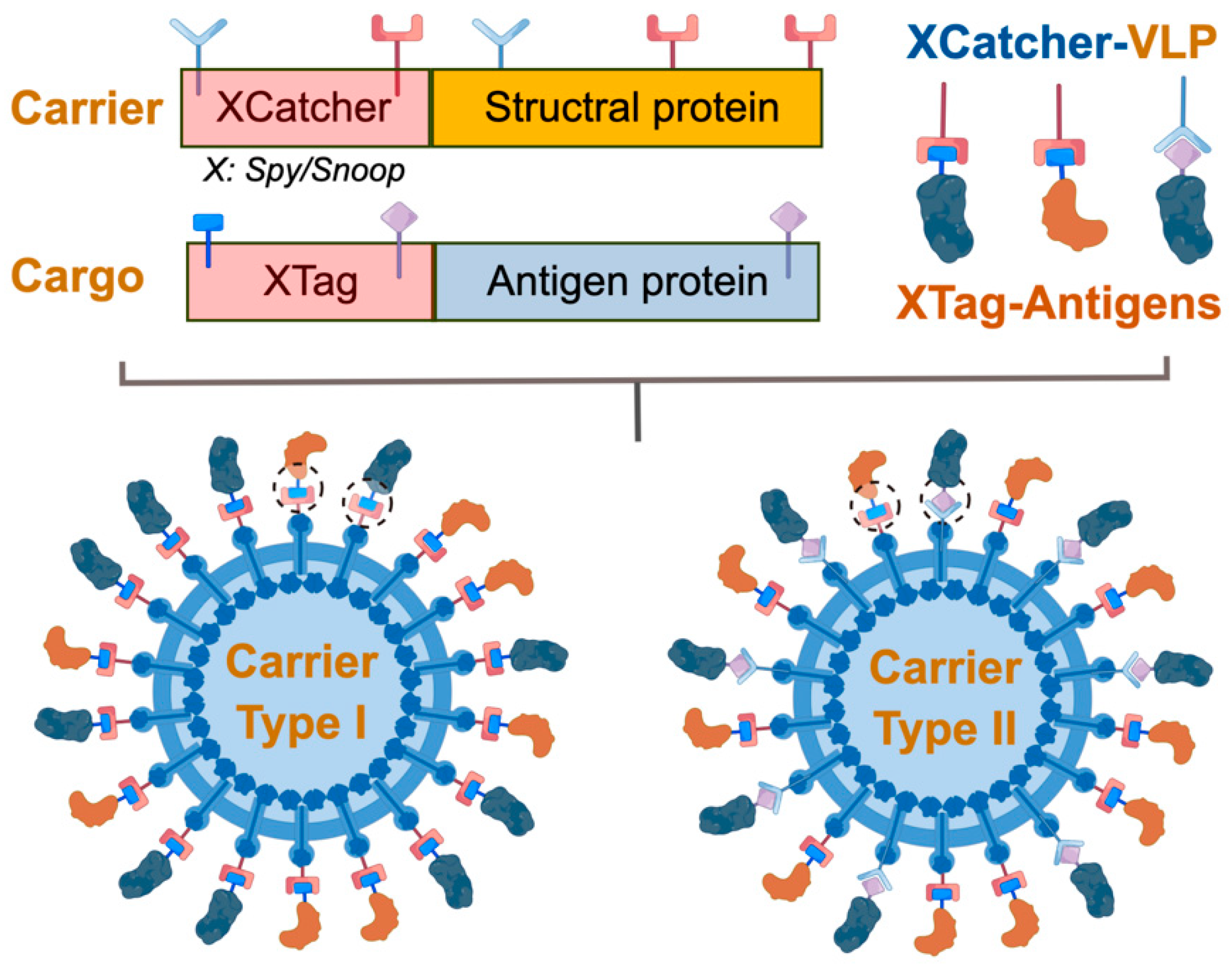
4. Plug-and-Display Technology Using Covalent Binding Partners for VLP- and Nanoparticle-Based Vaccines
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; Huang, D.Q.; Zou, B.; Yang, H.; Hui, W.Z.; Rui, F.; Yee, N.T.S.; Liu, C.; Nerurkar, S.N.; Kai, J.C.Y.; et al. Epidemiology of COVID-19: A Systematic Review and Meta-Analysis of Clinical Characteristics, Risk Factors, and Outcomes. J. Med. Virol. 2021, 93, 1449–1458. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 1449–1458. [Google Scholar] [CrossRef]
- COVID-19 Research and Innovation Achievements, April 2021. Available online: https://www.who.int/publictions/m/item/covid-19-research-and-innovation-achievements (accessed on 7 September 2023).
- Zeltins, A. Construction and Characterization of Virus-Like Particles: A Review. Mol. Biotechnol. 2013, 53, 727–733. [Google Scholar] [CrossRef]
- Donaldson, B.; Lateef, Z.; Walker, G.F.; Young, S.L.; Ward, V.K. Virus-Like Particle Vaccines: Immunology and Formulation for Clinical Translation. Expert Rev. Vaccines 2018, 17, 833–849. [Google Scholar] [CrossRef]
- Fuenmayor, J.; Godia, F.; Cervera, L. Production of Virus-Like Particles for Vaccines. N. Biotechnol. 2017, 39, 174–180. [Google Scholar] [CrossRef]
- Xu, J.; Hiramatsu, R.; Suhaimi, H.; Kato, T.; Fujimoto, A.; Tokiwa, T.; Ike, K.; Park, E.Y. Neospora Caninum Antigens Displaying Virus-Like Particles as A Bivalent Vaccine Candidate Against Neosporosis. Vaccine 2019, 37, 6426–6434. [Google Scholar] [CrossRef]
- Marini, A.; Zhou, Y.; Li, Y.; Taylor, I.J.; Leneghan, D.B.; Jin, J.; Zaric, M.; Mekhaiel, D.; Long, C.A.; Miura, K.; et al. A Universal Plug-and-Display Vaccine Carrier Based on HBsAg VLP to Maximize Effective Antibody Response. Front. Immunol. 2019, 10, 2931. [Google Scholar] [CrossRef]
- Barsoe, S.; Skovgaard, K.; Sepulveda, D.; Stratmann, A.; Vendramin, N.; Lorenzen, N. Nervous Necrosis Virus-like Particle (VLP) Vaccine Stimulates European Sea Bass Innate and Adaptive Immune Responses and Induces Long-Term Protection Against Disease. Pathogens 2021, 10, 1477. [Google Scholar] [CrossRef]
- Cimica, V.; Saleem, S.; Matuczinski, E.; Adams-Fish, D.; McMahon, C.; Rashid, S.; Stedman, T.T. A Virus-Like Particle-Based Vaccine Candidate Against the Tick-Borne Powassan Virus Induces Neutralizing Antibodies in a Mouse Model. Pathogens 2021, 10, 680. [Google Scholar] [CrossRef]
- Tariq, H.; Batool, S.; Asif, S.; Ali, M.; Abbasi, B.H. Virus-Like Particles: Revolutionary Platforms for Developing Vaccines Against Emerging Infectious Diseases. Front. Microbiol. 2022, 12, 790121. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-Ncov Spike in The Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional Assessment of Cell Entry and Receptor Usage For SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Shalash, A.O.; Hussein, W.M.; Skwarczynski, M.; Toth, I. Key Considerations for The Development of Safe and Effective SARS-Cov-2 Subunit Vaccine: A Peptide-Based Vaccine Alternative. Adv. Sci. 2021, 8, e2100985. [Google Scholar] [CrossRef] [PubMed]
- Yadav, T.; Kumar, S.; Mishra, G.; Saxena, S.K. Tracking the COVID-19 vaccines: The global landscape. Hum. Vaccin. Immunother. 2023, 19, 2191577. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-Like Particles: Preparation, Immunogenicity and Their Roles as Nanovaccines and Drug Nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Shi, M.; Li, J.; Song, P.; Li, N. Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol. 2020, 8, 862. [Google Scholar] [CrossRef]
- Ghorbani, A.; Zare, F.; Sazegari, S.; Afsharifar, A.; Eskandari, M.H.; Pormohammad, A. Development of a Novel Platform of Virus-Like Particle (VLP)-Based Vaccine Against COVID-19 by Exposing Epitopes: An Immunoinformatic Approach. New Microbes New Infect. 2020, 38, 100786. [Google Scholar] [CrossRef]
- Brouwer, P.J.M.; Brinkkemper, M.; Maisonnasse, P.; Dereuddre-Bosquet, N.; Grobben, M.; Claireaux, M.; de Gast, M.; Marlin, R.; Chesnais, V.; Diry, S.; et al. Two-Component Spike Nanoparticle Vaccine Protects Macaques From SARS-CoV-2 Infection. Cell 2021, 184, 1188–1200. [Google Scholar] [CrossRef]
- Deo, V.K.; Yoshimatsu, K.; Otsuki, T.; Dong, J.; Kato, T.; Park, E.Y. Display of Neospora Caninum Surface Protein Related Sequence 2 on Rous Sarcoma Virus-Derived Gag Protein Virus-Like Particles. J. Biotechnol. 2013, 165, 69–75. [Google Scholar] [CrossRef]
- Kato, T.; Takami, Y.; Kumar Deo, V.; Park, E.Y. Preparation of Virus-Like Particle Mimetic Nanovesicles Displaying the S Protein of Middle East Respiratory Syndrome Coronavirus Using Insect Cells. J. Biotechnol. 2019, 306, 177–184. [Google Scholar] [CrossRef]
- Utomo, D.I.S.; Pambudi, S.; Sjatha, F.; Kato, T.; Park, E.Y. Production of Dengue Virus-Like Particles Serotype-3 In Silkworm Larvae and Their Ability to Elicit a Humoral Immune Response in Mice. AMB Express 2020, 10, 147. [Google Scholar] [CrossRef] [PubMed]
- Gwon, Y.D.; Kim, S.; Cho, Y.; Heo, Y.; Cho, H.; Park, K.; Lee, H.J.; Choi, J.; Poo, H.; Kim, Y.B. Immunogenicity of Virus Like Particle Forming Baculoviral DNA Vaccine Against Pandemic Influenza H1N1. PLoS ONE 2016, 11, e0154824. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, Y.; Deo, V.K.; Kato, T.; Park, E.Y. Production of Rous Sarcoma Virus-Like Particles Displaying Human Transmembrane Protein in Silkworm Larvae and its Application to Ligand-Receptor Binding Assay. J. Biotechnol. 2011, 155, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Lua, L.H.; Connors, N.K.; Sainsbury, F.; Chuan, Y.P.; Wibowo, N.; Middelberg, A.P. Bioengineering Virus-Like Particles as Vaccines. Biotechnol. Bioeng. 2014, 111, 425–440. [Google Scholar] [CrossRef]
- Garg, H.; Mehmetoglu-Gurbuz, T.; Joshi, A. Virus Like Particles (VLP) as Multivalent Vaccine Candidate Against Chikungunya, Japanese Encephalitis, Yellow Fever and Zika Virus. Sci. Rep. 2020, 10, 4017. [Google Scholar] [CrossRef]
- Masuda, A.; Lee, J.; Miyata, T.; Mon, H.; Sato, K.; Oyama, K.; Sakurai, Y.; Yasuda, J.; Takahashi, D.; Ueda, T.; et al. Optimization of SARS-CoV-2 Spike Protein Expression in the Silkworm and Induction of Efficient Protective Immunity by Innoculation With Alum Adjuvants. Front. Immunol. 2022, 12, 803647. [Google Scholar] [CrossRef]
- Steele, K.H.; Stone, B.J.; Franklin, K.M.; Fath-Goodin, A.; Zhang, X.; Jiang, H.; Webb, B.A.; Geisler, C. Improving The Baculovirus Expression Vector System With Vankyrin-Enhanced Technology. Biotechnol. Prog. 2017, 33, 1496–1507. [Google Scholar] [CrossRef]
- Chambers, A.C.; Aksular, M.; Graves, L.P.; Irons, S.L.; Possee, R.D.; King, L.A. Overview of the Baculovirus Expression System. Curr. Protoc. Protein Sci. 2018, 91, 541–546. [Google Scholar] [CrossRef]
- Neuhold, J.; Radakovics, K.; Lehner, A.; Weissmann, F.; Garcia, M.Q.; Romero, M.C.; Berrow, N.S.; Stolt-Bergner, P. Goldenbac: A Simple, Highly Efficient, And Widely Applicable System for Construction of Multi-Gene Expression Vectors for Use with The Baculovirus Expression Vector System. BMC Biotechnol. 2020, 20, 26. [Google Scholar] [CrossRef]
- Berger, I.; Fitzgerald, D.J.; Richmond, T.J. Baculovirus Expression System for Heterologous Multiprotein Complexes. Nat. Biotechnol. 2004, 22, 1583–1587. [Google Scholar] [CrossRef]
- Hong, M.; Li, T.; Xue, W.; Zhang, S.; Cui, L.; Wang, H.; Zhang, Y.; Zhou, L.; Gu, Y.; Xia, N.; et al. Genetic Engineering of Baculovirus-Insect Cell System to Improve Protein Production. Front. Bioeng. Biotechnol. 2022, 10, 994743. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, F.; Teixeira, A.P.; Carinhas, N.; Carrondo, M.J.; Alves, P.M. Insect Cells as A Production Platform of Complex Virus-Like Particles. Expert Rev. Vaccines 2013, 12, 225–236. [Google Scholar] [CrossRef]
- Vicente, T.; Roldao, A.; Peixoto, C.; Carrondo, M.J.; Alves, P.M. Large-scale Production and Purification of VLP-Based Vaccines. J. Invertebr. Pathol. 2011, 107, S42–S48. [Google Scholar] [CrossRef]
- Kato, T.; Sugioka, S.; Itagaki, K.; Enoch, Y.P. Gene transduction in mammalian cells using Bombyx mori nucleopolyhedrovirus assisted by glycoprotein 64 of Autographa California multiple nucleopolyhedrovirus. Sci. Rep. 2016, 6, 32283. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Sekiguchi, T.; Boonyakida, J.; Kato, T.; Park, E.Y. Display of Multiple Proteins on Engineered Canine Parvovirus-like Particles Expressed in Cultured Silkworm Cells and Silkworm Larvae. Front. Bioeng. Biotechnol. 2023, 11, 1096363. [Google Scholar] [CrossRef] [PubMed]
- Felberbaum, R.S. The Baculovirus Expression Vector System: A Commercial Manufacturing Platform for Viral Vaccines and Gene Therapy Vectors. Biotechnol. J. 2015, 10, 702–714. [Google Scholar] [CrossRef]
- Kato, T.; Arai, S.; Ichikawa, H.; Park, E.Y. Versatility of Chitosan/BmNPV Bacmid DNA Nanocomplex As Transfection Reagent of Recombinant Protein Expression in Silkworm Larvae. Biotechnol. Lett. 2016, 38, 1449–1457. [Google Scholar] [CrossRef]
- Fujita, R.; Hino, M.; Ebihara, T.; Nagasato, T.; Masuda, A.; Lee, J.M.; Fujii, T.; Mon, H.; Kakino, K.; Nagai, R.; et al. Efficient Production of Recombinant SARS-Cov-2 Spike Protein Using the Baculovirus-Silkworm System. Biochem. Biophys. Res. Commun. 2020, 529, 257–262. [Google Scholar] [CrossRef]
- Tomita, M. Development of Large-Scale Silkworm-rearing Technologies for the GMP Production of Biologics. Yakugaku Zasshi 2018, 138, 875–884. [Google Scholar] [CrossRef]
- Laurens, M.B. RTS, S/AS01 Vaccine (Mosquirix™): An Overview. Hum. Vaccin Immunother. 2020, 16, 480–489. [Google Scholar] [CrossRef]
- Ye, Q.; Lin, X.; Wang, T.; Cui, Y.; Jiang, H.; Lu, Y. Programmable Protein Topology Via Spycatcher-Spytag Chemistry in One-Pot Cell-Free Expression System. Protein Sci. 2022, 31, e4355. [Google Scholar] [CrossRef] [PubMed]
- Urakami, A.; Ngwe Tun, M.M.; Moi, M.L.; Sakurai, A.; Ishikawa, M.; Kuno, S.; Ueno, R.; Morita, K.; Akahata, W. An Envelope-Modified Tetravalent Dengue Virus-Like-Particle Vaccine has Implications for Flavivirus Vaccine Design. J. Virol. 2017, 91, e01181-17. [Google Scholar] [CrossRef] [PubMed]
- Utomo, D.I.S.; Pambudi, S.; Park, E.Y. Humoral Immune Response Induced with Dengue Virus-Like Particles Serotypes 1 and 4 Produced in Silkworm. AMB Express 2022, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Peabody, J.; Pang, Y.S.; Schiller, J.; Chackerian, B.; Tumban, E. A Novel Candidate HPV Vaccine: MS2 Phage VLP Displaying a Tandem HPV L2 Peptide Offers Similar Protection in Mice To Gardasil-9. Antivir. Res. 2017, 147, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, A.L.; Tagliamonte, M.; Buonaguro, F.M.; Tornesello, M.L.; Buonaguro, L. Virus-like Particles as Preventive and Therapeutic Cancer Vaccines. Vaccines. 2022, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Cheng, K. The Principles and Applications of Avidin-based Nanoparticles in Drug Delivery and Diagnosis. J. Control. Release 2017, 245, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Lizotte, P.H.; Wen, A.M.; Sheen, M.R.; Fields, J.; Rojanasopondist, P.; Steinmetz, N.F.; Fiering, S. In Situ Vaccination with Cowpea Mosaic Virus Nanoparticles Suppresses Metastatic Cancer. Nat. Nanotechnol. 2016, 11, 295–303. [Google Scholar] [CrossRef]
- Brune, K.D.; Howarth, M. New Routes and Opportunities for Modular Construction of Particulate Vaccines: Stick, Click, and Glue. Front. Immunol. 2018, 9, 1432. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Augusto, G.; Bachmann, M.F. The 3Ds in Virus-Like Particle Based-Vaccines: “Design, Delivery and Dynamics”. Immunol. Rev. 2020, 296, 155–168. [Google Scholar] [CrossRef]
- Juraszek, J.; Rutten, L.; Blokland, S.; Bouchier, P.; Voorzaat, R.; Ritschel, T.; Bakkers, M.J.G.; Renault, L.R.L.; Langedijk, J.P.M. Stabilizing the closed SARS-CoV-2 spike trimer. Nat. Commun. 2021, 12, 244. [Google Scholar] [CrossRef]
- Riley, T.P.; Chou, H.-T.; Hu, R.; Bzymek, K.P.; Correia, A.R.; Partin, A.C.; Li, D.; Gong, D.; Wang, Z.; Yu, X.; et al. Enhancing the Prefusion Conformational Stability of SARS-CoV-2 Spike Protein Through Structure-Guided Design. Front. Immunol. 2021, 12, 660198. [Google Scholar] [CrossRef]
- Hsieh, C.-L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.-C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, I.C.; Ipekoglu, E.M.; Bulbul, A.; Turay, N.; Yildirim, M.; Evcili, I.; Yilmaz, N.S.; Guvencli, N.; Aydin, Y.; Gungor, B.; et al. Development and Preclinical Evaluation of Virus-Like Particle Vaccine Against COVID-19 Infection. Allergy 2022, 77, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Haynes, W.A.; Kamath, K.; Bozekowski, J.; Baum-Jones, E.; Campbell, M.; Casanovas-Massana, A.; Daugherty, P.S.; Cruz, C.S.D.; Dhal, A.; Farhadian, S.F.; et al. High-Resolution Epitope Mapping and Characterization of SARS-CoV-2 Antibodies in Large Cohorts of Subjects With COVID-19. Commun. Biol. 2021, 4, 1317. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Kode, V.; Bhojak, K.; Karunakaran, C.; Lee, K.; Manoharan, M.; Ramesh, A.; Hv, S.; Srivastava, A.; Sathian, R.; et al. Immunodominant T-cell epitopes from the SARS-CoV-2 spike antigen reveal robust pre-existing T-cell immunity in unexposed individuals. Sci. Rep. 2021, 11, 13164. [Google Scholar] [CrossRef]
- Swann, H.; Sharma, A.; Preece, B.; Peterson, A.; Eldredge, C.; Belnap, D.M.; Vershinin, M.; Saffarian, S. Minimal System for Assembly of SARS-CoV-2 Virus Like Particles. Sci. Rep. 2020, 10, 21877. [Google Scholar] [CrossRef]
- Mi, Y.; Xie, T.; Zhu, B.; Tan, J.; Li, X.; Luo, Y.; Li, F.; Niu, H.; Han, J.; Lv, W.; et al. Production of SARS-CoV-2 Virus-Like Particles in Insect Cells. Vaccines 2021, 9, 554. [Google Scholar] [CrossRef]
- Hennrich, A.A.; Sawatsky, B.; Santos-Mandujano, R.; Banda, D.H.; Oberhuber, M.; Schopf, A.; Pfaffinger, V.; Wittwer, K.; Riedel, C.; Pfaller, C.K.; et al. Safe and Effective Two-In-One Replicon-and-VLP Minispike Vaccine For COVID-19: Protection of Mice After a Single Immunization. PLoS Pathog. 2021, 17, e1009064. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, W.; Abiona, O.M.; Nazzari, A.; Olia, A.S.; Ou, L.; Phung, E.; Stephens, T.; Tsybovsky, Y.; Verardi, R.; et al. Newcastle Disease Virus-Like Particles Displaying Prefusion-Stabilized SARS-CoV-2 Spikes Elicit Potent Neutralizing Responses. Vaccines 2021, 9, 73. [Google Scholar] [CrossRef]
- Liu, X.; Chang, X.; Rothen, D.; Derveni, M.; Krenger, P.; Roongta, S.; Wright, E.; Vogel, M.; Tars, K.; Mohsen, M.O.; et al. AP205 VLPs Based on Dimerized Capsid Proteins Accommodate RBM Domain of SARS-CoV-2 and Serve as an Attractive Vaccine Candidate. Vaccines 2021, 9, 403. [Google Scholar] [CrossRef]
- Carter, D.C.; Wright, B.; Jerome, W.G.; Rose, J.P.; Wilson, E. A Unique Protein Self-Assembling Nanoparticle with Significant Advantages in Vaccine Development and Production. J. Nanomater. 2020, 2020, 4297937. [Google Scholar] [CrossRef]
- Thrane, S.; Janitzek, C.M.; Agerbæk, M.; Ditlev, S.B.; Resende, M.; Nielsen, M.A.; Theander, T.G.; Salanti, A.; Sander, A.F. A Novel Virus-Like Particle Based Vaccine Platform Displaying the Placental Malaria Antigen VAR2CSA. PLoS ONE 2015, 10, e0143071. [Google Scholar] [CrossRef] [PubMed]
- Tumban, E.; Peabody, J.; Tyler, M.; Peabody, D.S.; Chackerian, B. VLPs Displaying a Single L2 Epitope Induce Broadly Cross-Neutralizing Antibodies against Human Papillomavirus. PLoS ONE 2012, 7, e49751. [Google Scholar] [CrossRef]
- Chiba, S.; Frey, S.J.; Halfmann, P.J.; Kuroda, M.; Maemura, T.; Yang, J.E.; Wright, E.R.; Kawaoka, Y.; Kane, R.S. Multivalent Nanoparticle-Based Vaccines Protect Hamsters Against SARS-CoV-2 After A Single Immunization. Commun. Biol. 2021, 4, 597. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide Tag Forming a Rapid Covalent Bond to a Protein, Through Engineering a Bacterial Adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef] [PubMed]
- Reddington, S.C.; Howarth, M. Secrets of a Covalent Interaction for Biomaterials and Biotechnology: SpyTag and SpyCatcher. Curr Opin Chem Biol. 2015, 29, 94–99. [Google Scholar] [CrossRef]
- Li, L.; Fierer, J.O.; Rapoport, T.A.; Howarth, M. Structural Analysis and Optimization of The Covalent Association Between Spycatcher and A Peptide Tag. J. Mol. Biol. 2014, 426, 309–317. [Google Scholar] [CrossRef]
- Brune, K.D.; Leneghan, D.B.; Brian, I.J.; Ishizuka, A.S.; Bachmann, M.F.; Draper, S.J.; Biswas, S.; Howarth, M. Plug-and-Display: Decoration of Virus-Like Particles via Isopeptide Bonds for Modular Immunization. Sci. Rep. 2016, 6, 19234. [Google Scholar] [CrossRef]
- Xu, J.; Kato, T.; Park, E.Y. Development of SpyTag/SpyCatcher-Bacmid Expression Vector System (SpyBEVS) for Protein Bioconjugations Inside of Silkworms. Int. J. Mol. Sci. 2019, 20, 4228. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.X.; Zhang, D.; Wu, S.; Zhang, G. Enhanced Thermal Stability of Lichenase from Bacillus Subtilis 168 By Spytag/Spycatcher-Mediated Spontaneous Cyclization. Biotechnol. Biofuels 2016, 9, 79. [Google Scholar] [CrossRef]
- Sharma, A.; Preece, B.; Swann, H.; Fan, X.; McKenney, R.J.; Ori-McKenney, K.M.; Saffarian, S.; Vershinin, M.D. Structural Stability of SARS-CoV-2 Virus Like Particles Degrades with Temperature. Biochem. Biophys. Res. Commun. 2021, 534, 343–346. [Google Scholar] [CrossRef]
- Rahikainen, R.; Rijal, P.; Tan, T.K.; Wu, H.J.; Andersson, A.C.; Barrett, J.R.; Bowden, T.A.; Draper, S.J.; Townsend, A.R.; Howarth, M. Overcoming Symmetry Mismatch in Vaccine Nanoassembly through Spontaneous Amidation. Angew. Chem. Int. Ed. Engl. 2021, 60, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Veggiani, G.; Nakamura, T.; Brenner, M.D.; Gayet, R.V.; Yan, J.; Robinson, C.V.; Howarth, M. Programmable Polyproteams Built Using Twin Peptide Superglues. Proc. Natl. Acad. Sci. USA 2016, 113, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, K.; Aoki, W.; Koike, N.; Ueda, M. Development of a Yeast Cell Surface Display Method Using the Spytag/Spycatcher System. Sci. Rep. 2021, 11, 11059. [Google Scholar] [CrossRef]
- Stander, J.; Chabeda, A.; Rybicki, E.P.; Meyers, A.E. A Plant-Produced Virus-Like Particle Displaying Envelope Protein Domain III Elicits an Immune Response Against West Nile Virus in Mice. Front. Plant. Sci. 2021, 12, 738619. [Google Scholar] [CrossRef]
- Gallus, S.; Mittmann, E.; Rabe, K.S. A Modular System for the Rapid Comparison of Different Membrane Anchors for Surface Display on Escherichia coli. Chembiochem 2022, 2, e202100472. [Google Scholar] [CrossRef]
- Shrocket, E.; Fujimura, E.; Kula, T.; Timms, R.T.; Lee, I.H.; Leng, Y.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; Chen, Y.; et al. Viral Epitope Profiling of COVID-19 Patients Reveals Cross-reactivity And Correlates of Severity. Science 2020, 370, eabd4250. [Google Scholar] [CrossRef]
- Vragniau, C.; Bufton, J.C.; Garzoni, F.; Stermann, E.; Rabi, F.; Terrat, C.; Guidetti, M.; Josserand, V.; Williams, M.; Woods, C.J.; et al. Synthetic Self-Assembling Addomer Platform for Highly Efficient Vaccination By Genetically Encoded Multiepitope Display. Sci. Adv. 2019, 5, eaaw2853. [Google Scholar] [CrossRef]
- Chevillard, C.; Amen, A.; Besson, S.; Hannani, D.; Bally, I.; Dettling, V.; Gout, E.; Moreau, C.J.; Buisson, M.; Gallet, S.; et al. Elicitation of Potent SARS-CoV-2 Neutralizing Antibody Responses Through Immunization with a Versatile Adenovirus-Inspired Multimerization Platform. Mol. Ther. 2022, 30, 1913–1925. [Google Scholar] [CrossRef]
- Ma, X.; Zou, F.; Yu, F.; Li, R.; Yuan, Y.; Zhang, Y.; Zhang, X.; Deng, J.; Chen, T.; Song, Z.; et al. Nanoparticle Vaccines Based on the Receptor Binding Domain (RBD) and Heptad Repeat (HR) of SARS-CoV-2 Elicit Robust Protective Immune Responses. Immunity 2020, 53, 1315–1330. [Google Scholar] [CrossRef]
- Zhang, B.; Chao, C.W.; Tsybovsky, Y.; Abiona, O.M.; Hutchinson, G.B.; Moliva, J.I.; Olia, A.S.; Pegu, A.; Phung, E.; Stewart-Jones, G.B.E.; et al. A platform Incorporating Trimeric Antigens into Self-Assembling Nanoparticles Reveals SARS-CoV-2-Spike Nanoparticles to Elicit Substantially Higher Neutralizing Responses Than Spike Alone. Sci. Rep. 2020, 10, 18149. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.K.; Rijal, P.; Rahikainen, R.; Keeble, A.H.; Schimanski, L.; Hussain, S.; Harvey, R.; Hayes, J.W.P.; Edwards, J.C.; McLean, R.K.; et al. A COVID-19 Vaccine Candidate Using SpyCatcher Multimerization of the SARS-CoV-2 Spike Protein Receptor-Binding Domain Induces Potent Neutralising Antibody Responses. Nat. Commun. 2021, 12, 542. [Google Scholar] [CrossRef]
- Wang, W.; Huang, B.; Zhu, Y.; Tan, W.; Zhu, M. Ferritin Nanoparticle-Based SARS-CoV-2 RBD Vaccine Induces a Persistent Antibody Response and Long-Term Memory in Mice. Cell. Mol. Immunol. 2021, 18, 749–751. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, Y.; Wu, B.; Wang, X.; Lin, Y.; Luo, Y.; Li, R.; Chen, T.; Deng, J.; Zhang, X.; et al. Improvement of a SARS-CoV-2 vaccine by Enhancing the Conjugation Efficiency of The Immunogen to Self-Assembled Nanoparticles. Cell. Mol. Immunol. 2021, 18, 2042–2044. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.F.; Sun, C.; Zhuang, Z.; Yuan, R.Y.; Zheng, Q.; Li, J.P.; Zhou, P.P.; Chen, X.C.; Liu, Z.; Zhang, X.; et al. Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. ACS Nano 2021, 15, 2738–2752. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Lin, X.; Wang, Y.; Abraham, C.; Sou, C.; Ngo, T.; Zhang, Y.; Wilson, I.A.; Zhu, J. Single-Component, Self-Assembling, Protein Nanoparticles Presenting the Receptor Binding Domain and Stabilized Spike As SARS-CoV-2 Vaccine Candidates. Sci. Adv. 2021, 7, eabf1591. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Castro, A.; Loeffler, K.; Frey, S.J.; Chiba, S.; Kawaoka, Y.; Kane, R.S. Potent Neutralization Of SARS-CoV-2 Including Variants of Concern by Vaccines Presenting the Receptor-Binding Domain Multivalently from Nanoscaffolds. Bioeng. Transl. Med. 2021, 6, e10253. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; Gnanapragasam, P.N.P.; Lee, Y.E.; Hoffman, P.R.; Ou, S.; Kakutani, L.M.; Keeffe, J.R.; Wu, H.J.; Howarth, M.; West, A.P.; et al. Mosaic Nanoparticles Elicit Cross-Reactive Immune Responses to Zoonotic Coronaviruses in Mice. Science 2021, 371, 735–741. [Google Scholar] [CrossRef]
- Lu, S.; Xie, X.X.; Zhao, L.; Wang, B.; Zhu, J.; Yang, T.R.; Yang, G.W.; Ji, M.; Lv, C.P.; Xue, J.; et al. The Immunodominant and Neutralization Linear Epitopes For SARS-CoV-2. Cell Rep. 2021, 34, 108666. [Google Scholar] [CrossRef]
- Guo, Y.; He, W.; Mou, H.; Zhang, L.; Chang, J.; Peng, S.; Ojha, A.; Tavora, R.; Parcells, M.S.; Luo, G.; et al. An Engineered Receptor-Binding Domain Improves the Immunogenicity of Multivalent SARS-CoV-2 Vaccines. Mbio 2021, 12, 10–1128. [Google Scholar] [CrossRef]
- World Health Organization. COVID-19—Landscape of Novel Coronavirus Candidate Vaccine Development Worldwide. COVID-19 Vaccine Tracker and Landscape. 2023. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 7 September 2023).
- Sharifzadeh, M.; Mottaghi-Dastjerdi, N.; Soltany Rezae Raad, M. A Review of Virus-Like Particle-Based SARS-CoV-2 Vaccines in Clinical Trial Phases. Iran. J. Pharm. Res. 2022, 21, e127042. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.B.; Freire, J.M.; Moleirinho, M.G.; Monteiro, F.; Gaspar, D.; Castanho, M.A.R.B.; Carrondo, M.J.T.; Alves, P.M.; Bernardes, G.J.L.; Peixoto, C. Bioorthogonal Strategy for Bioprocessing of Specific-Site-Functionalized Enveloped Influenza-Virus-Like Particles. Bioconjug Chem. 2016, 27, 2386–2399. [Google Scholar] [CrossRef] [PubMed]
- Cervera, L.; Gòdia, F.; Tarrés-Freixas, F.; Aguilar-Gurrieri, C.; Carrillo, J.; Blanco, J.; Gutiérrez-Granados, S. Production of HIV-1-based virus-like particles for vaccination: Achievements and limits. Appl. Microbiol. Biotechnol. 2019, 103, 7367–7384. [Google Scholar] [CrossRef] [PubMed]

| VLP Model | SARS-CoV-2 Antigen | Results | References |
|---|---|---|---|
| SARS-CoV-2 | Spike protein (S), membrane protein (M), and small envelope protein (E) | SARS-CoV-VLPs demonstrate molecular and morphological properties of native VLP | [17,57,58] |
| Vesicular stomatitis virus (VSVG/VSV∆G) | Chimeric spike protein receptor-binding domain (RBD) (Rhabdovirus RBD-miniSpikeminispike) | Mice immunization and its neutralizing antibodies: A single dose of VSVG/VSV∆G-miniSpikeminispike-eGFP (G) stimulates high titers of SARS-CoV-2 and protects transgenic K18-hACE2 mice from SARS-CoV-2. Homologous boost immunization of VSVG/VSV∆G-miniSpikeminispike-eGFP (G) enhanced the neutralizing antibody activity. | [59] |
| Newcastle disease virus (NDVLP) | Spike ectodomain (S2P) | Mice immunization and its neutralizing antibodies: S2P-NDVLP showed high spike-specific IgG titers and elicited a high humoral immune response. S2P-NDVLP can elicit substantial neutralizing activities. | [60] |
| Acinetobacter phage AP205 | Spike protein receptor-binding domain (RBD) | Mice immunization and its neutralizing antibodies: Induce high-level IgG antibodies and are able to neutralize wild type of SARS-CoV-2. | [61] |
| Spike (S) protein (unmodified and modified versions), membrane glycoprotein (M), envelope (E), and nucleocapsid (N) | Spike protein (S), membrane protein (M), and small envelope protein (E) | 2P with two proline substitutions (K986P and V987P), or 6P with six proline substitutions (F817P, A 892P, A899P, A942P, K986P, and V987P) to stabilize the prefusion conformation. Mice, rats, and ferrets immunization and its neutralizing antibodies: VLPs expressing the hexaproline stabilized prefusion spike antigen with Alum plus K3 CpG ODN adjuvant elicited high levels of anti-S, anti-RBD, anti-N IgG, and live virus-neutralizing antibodies. | [54] |
| VLP Model | SARS-CoV-2 Antigen | Results | References |
|---|---|---|---|
| SpyTag-RBD/SpyTag-HR; SpyCatcher-Ferritin “RBD/HR-SpyVLP” | Spike glycoprotein receptor-binding domain (RBD)/ Heptad repeat (HR) | Mice and rhesus macaques immunization, and its neutralizing antibodies: RBD/HR-SpyVLP vaccinated hACE2 transgenic mice reduced the viral load in the lung after challenge and promoted neutralizing antibodies and cellular immune response. RBD/HR vaccinated rhesus macaques induced T and B cell responses prior to boost immunization and neutralizing antibodies. | [81] |
| SpyTag-LUS/ Ferritin with addition N-linked glycan: SpyCatcher-SARS-CoV-2 Spike “SARS-CoV-2 spike-LUS nanoparticles” | Spike glycoprotein | Mice immunization and its neutralizing antibodies: SARS-CoV-2spike-LUS nanoparticles significantly improved its immunogenicity with a lower dose of immunogen and elicited a potent neutralization response compared with trimeric form. | [82] |
| SpyTag-RBD: SpyCatcher003-mi3 “RBD-SpyVLP” | Spike glycoprotein receptor-binding domain (RBD) | Mice and pig immunization, and its neutralizing antibodies: RBD-SpyVLP induces strong ACE2-blocking, highly immunogenic, and neutralizing antibodies response in mice and pig models. | [83] |
| SpyTag-RBD: SpyCatcher-Ferritin “Ferritin-NP-RBD” | Spike glycoprotein receptor-binding domain (RBD) | Mice immunization and its neutralizing antibodies: Ferritin-NP-RBD enhanced and gave persistent RBD-specific antibody response. Ferritin-NP-RBD had a neutralizing effect at serum dilution and showed inhibition to the infection in vitro SARS-CoV-2. | [84] |
| GvTagOpti-RBD: SdCatcher-HPF “RBD-Gv/Sd NPs” | Spike glycoprotein receptor-binding domain (RBD) | Mice immunization and its neutralizing antibodies: RBD-Gv/Sd NPs give higher titers of RBD-specific immune response and neutralizing antibodies. | [85] |
| SpyTag-RBD: ∆N1-SpyCatcher-mi3/Ferritin/I53-50 “RBD-NPs” | Spike glycoprotein receptor-binding domain (RBD) | Mice immunization and its neutralizing antibodies: RBD-NPs successfully block the binding of RBD to ACE2 and neutralize the antibody in vitro. | [86] |
| SpyTag-RBD/S2∆GHR2: SpyCatcher- Ferritin SApNP/I3-01v9 SApNP (Locking domain and T-helper epitope within the SApNP “RBD-Ferritin SApNP/S2∆GHR2-I3-01v9 SApNP” | Spike glycoprotein receptor-binding domain (RBD)/ Heptad repeat-deleted glycine-capped spike(S2∆GHR2) | Mice immunization and its neutralizing antibodies: RBD-Ferritin SApNP, S2∆GHR2-NPs induced more potent neutralizing antibody compared to the control. The S2∆GHR2-NPs critically elicited required T cell immunity. | [87] |
| SpyTag-RBD: SpyCatcher-mi3 “RBD-SpyCatcher-mi3” | Spike glycoprotein receptor-binding domain (RBD) | Mice immunization and its neutralizing antibodies: RBD-SpyCatcher-mi3 elicited a high neutralizing antibody titer against four variants, including the delta variant and isolated SARS-CoV-2. | [88] |
| SpyTag003-RBD/mosaic-4a/mosaic-4b/mosaic-8: SpyCatcher003-mi3 “RBD/mosaicRBD-NPs” | Spike glycoprotein receptor-binding domain (RBD), RBD from animal betacoronaviruses | Mice immunization and its neutralizing antibodies: RBD-NPs induced cross-reactive binding and neutralizing responses, whereas mosaicRBD-NPs induced antibodies with superior cross-reactive recognition of heterologous RBDs, of either RBD-NPs or COVID-19 convalescent human plasma. | [89] |
| SpyTag-S/E/M/N: SpyCatcher-HBc “HBc-S/E/M/N-P-VLPs” | B cell epitopes on spike (S), envelope (E), membrane (M), nucleocapsid (N) | Mice immunization and its neutralizing antibodies: HBc-S/E/M/N-P-VLPs showed the predicted epitopes from S/E/M/N that provide wide neutralizing and different immunodominant epitopes in mice models. | [90] |
| SpyTag-RBD-KLH/ S-KLH: SpyCatcher-mi3 60 “RBD-mi3/S-mi3” | Spike glycoprotein receptor-binding domain (RBD) and proline-stabilized S-protein ectodomain (S) | Rodent immunization and its neutralizing antibodies: RBD-mi3 is more immunogenic and elicits potent neutralizing responses compared to S-mi3 * Glycan-modification of RBD (gRBD) has been introduced and elicits a more potent neutralizing response than RBD. | [91] |
| ADDomer-SpyTag: RBD-SpyCatcher “ADD-RBD” | Spike glycoprotein receptor-binding domain (RBD) | Mice immunization and its neutralizing antibodies: A multimerized RBD display led to a strong improvement in immunogenicity and elicited a higher titer of neutralizing antibodies. | [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Setyo Utomo, D.I.; Suhaimi, H.; Muhammad Azami, N.A.; Azmi, F.; Mohd Amin, M.C.I.; Xu, J. An Overview of Recent Developments in the Application of Antigen Displaying Vaccine Platforms: Hints for Future SARS-CoV-2 VLP Vaccines. Vaccines 2023, 11, 1506. https://doi.org/10.3390/vaccines11091506
Setyo Utomo DI, Suhaimi H, Muhammad Azami NA, Azmi F, Mohd Amin MCI, Xu J. An Overview of Recent Developments in the Application of Antigen Displaying Vaccine Platforms: Hints for Future SARS-CoV-2 VLP Vaccines. Vaccines. 2023; 11(9):1506. https://doi.org/10.3390/vaccines11091506
Chicago/Turabian StyleSetyo Utomo, Doddy Irawan, Hamizah Suhaimi, Nor Azila Muhammad Azami, Fazren Azmi, Mohd Cairul Iqbal Mohd Amin, and Jian Xu. 2023. "An Overview of Recent Developments in the Application of Antigen Displaying Vaccine Platforms: Hints for Future SARS-CoV-2 VLP Vaccines" Vaccines 11, no. 9: 1506. https://doi.org/10.3390/vaccines11091506





