Study of the Effects of Several SARS-CoV-2 Structural Proteins on Antiviral Immunity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Viruses and Vaccines
2.2. Animal and Ethics
2.3. Plasmid Construction and Transfection
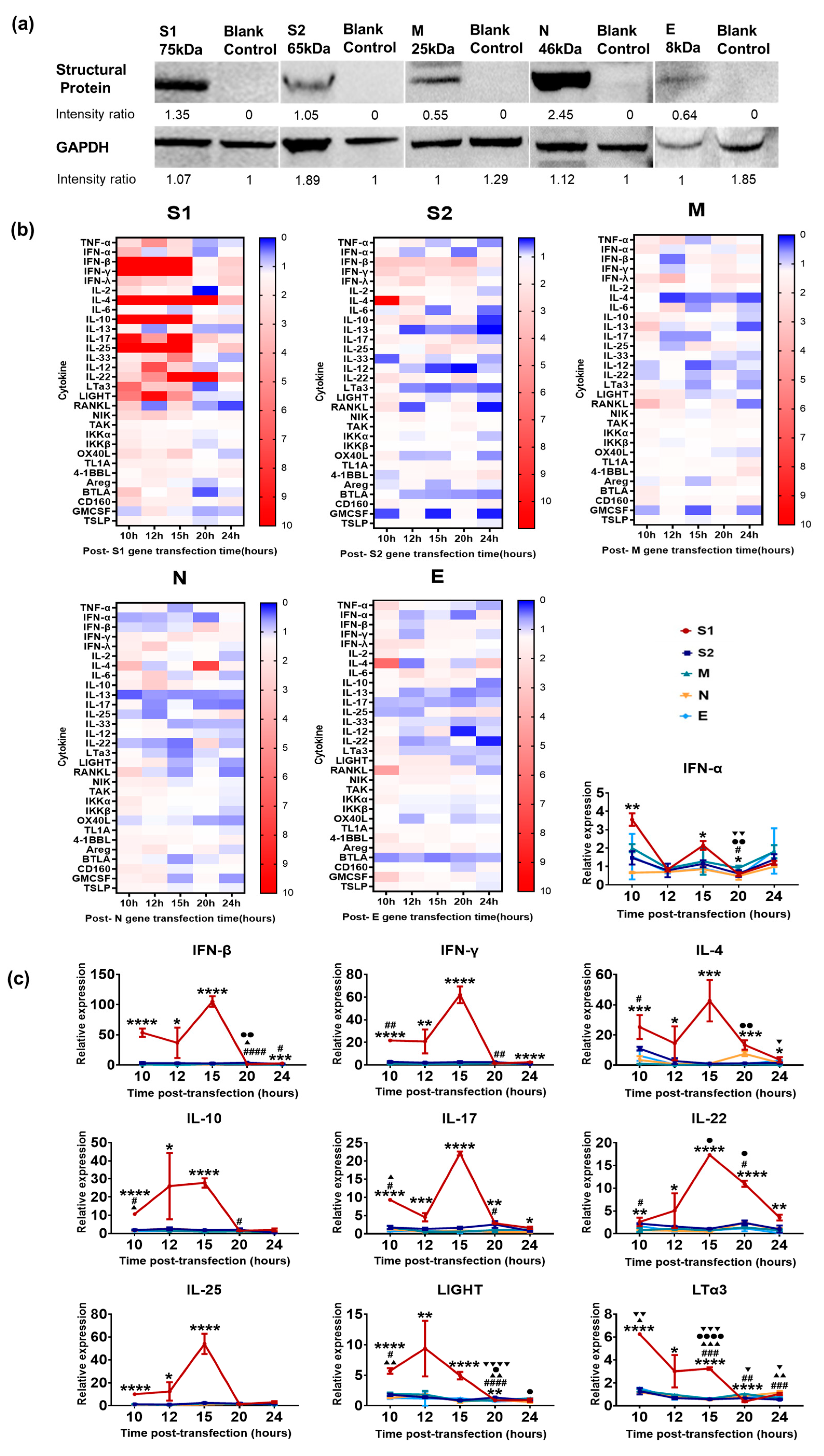
2.4. Western Blots
2.5. Cytokine Analysis (q-RT-PCR)
2.6. Animal Vaccination Protocol
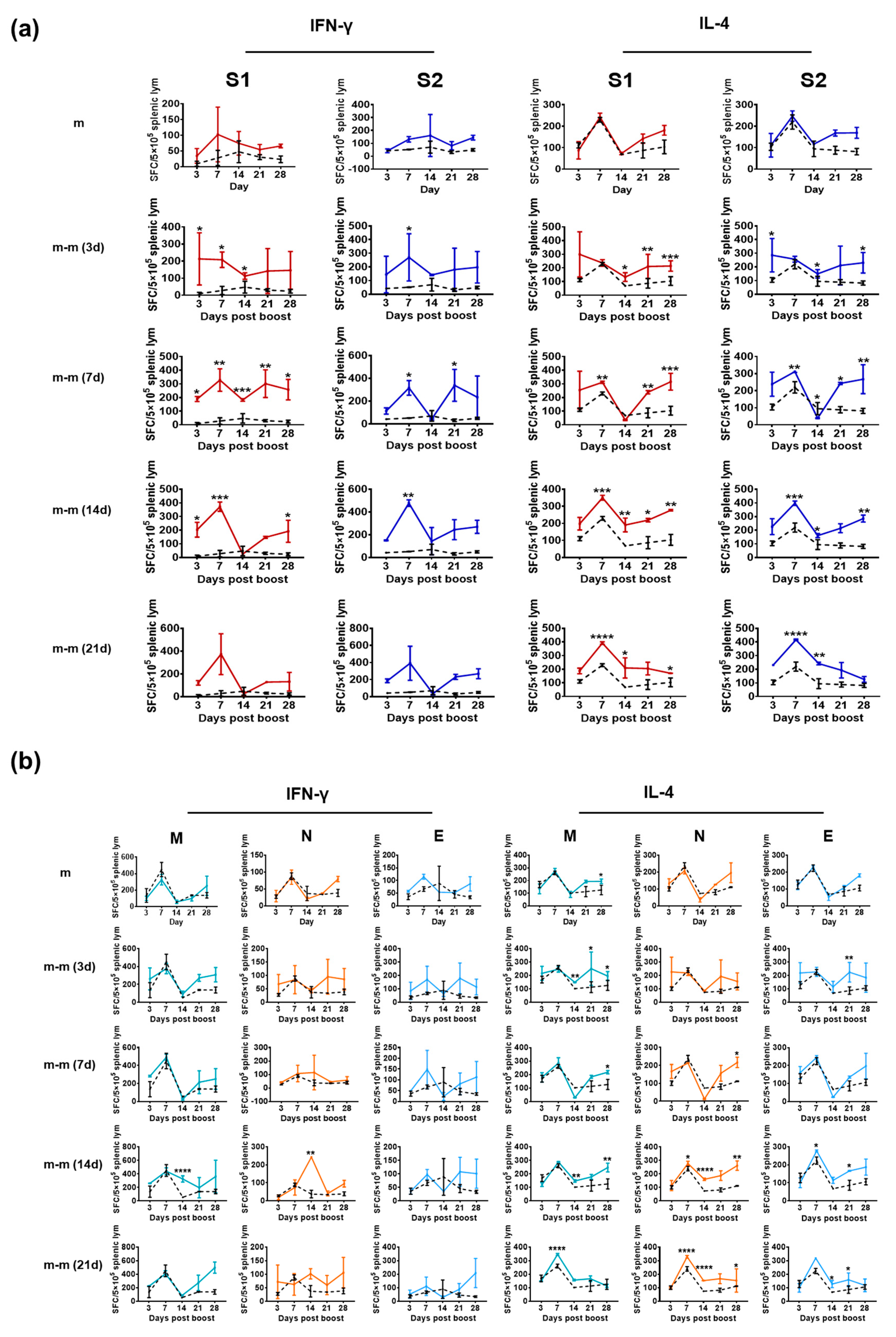
2.7. ELISpot Assay
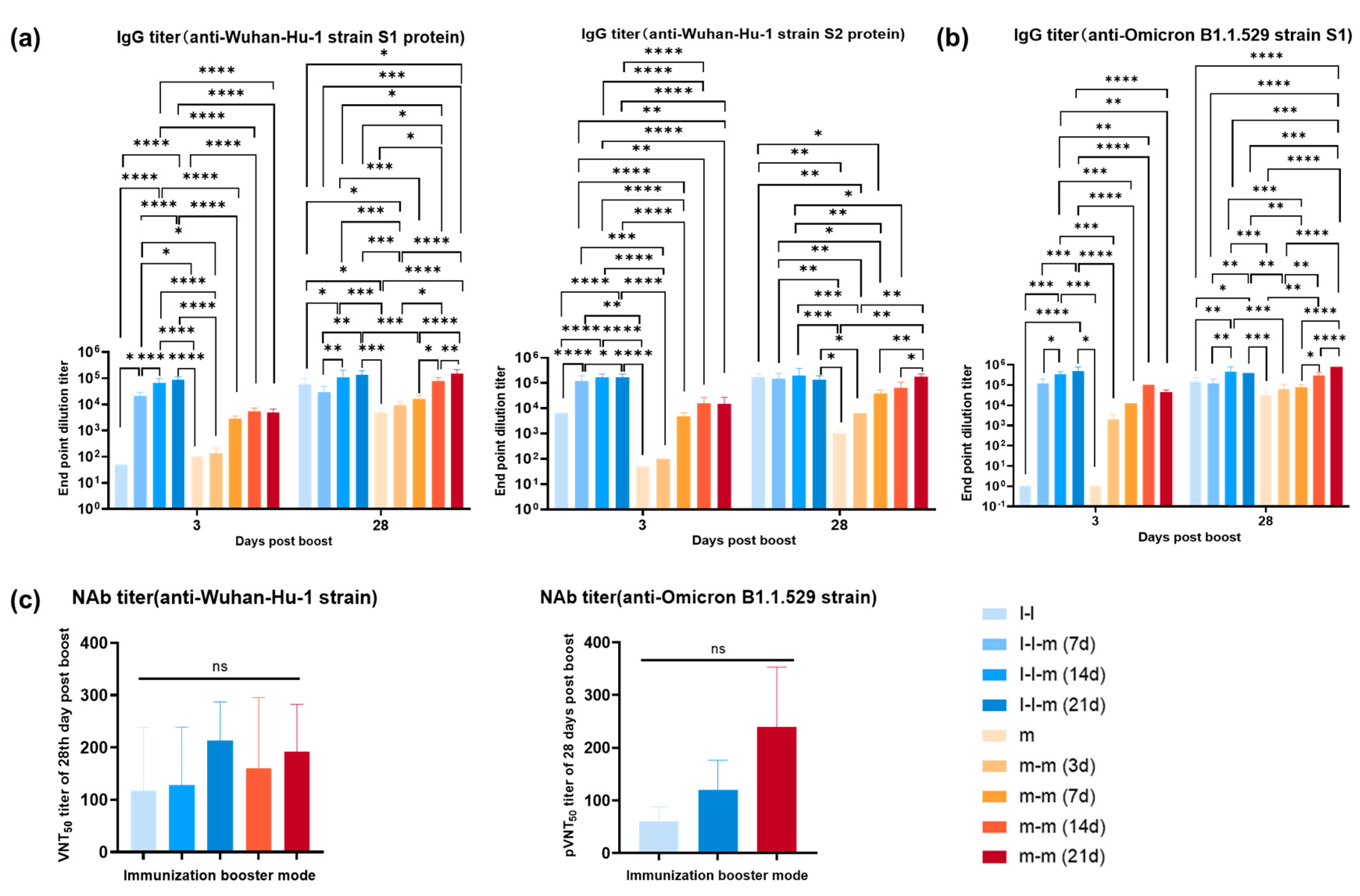
2.8. ELISA
2.9. Neutralizing Antibody Assay
2.10. Statistical Analysis
3. Results
3.1. Innate Immune Response Elicited by Various SARS-CoV-2 Structural Proteins in Epithelial Cells
3.2. Structural Proteins Elicit a Specific T-Cell Response in Mice Immunized with Inactivated SARS-CoV-2 Vaccine
3.3. Immunization with mRNA Vaccine for the S Gene Induces a Specific T-Cell Response against Only the S1 and S2 Proteins
3.4. Antibody Response Elicited by Two Doses of mRNA Vaccine or Two Doses of Inactivated Vaccine Followed by an mRNA Vaccine Boost in Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Walker, P.G.T.; Whittaker, C.; Watson, O.J.; Baguelin, M.; Winskill, P.; Hamlet, A.; Djafaara, B.A.; Cucunubá, Z.; Mesa, D.O.; Green, W.; et al. The impact of COVID-19 and strategies for mitigation and suppression in low- and middle-income countries. Science 2020, 369, 413–422. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Zhang, Z.; Mateus, J.; Coelho, C.H.; Dan, J.M.; Moderbacher, C.R.; Gálvez, R.I.; Cortes, F.H.; Grifoni, A.; Tarke, A.; Chang, J.; et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell 2022, 185, 2434–2451.e17. [Google Scholar] [CrossRef]
- Bai, Z.; Cao, Y.; Liu, W.; Li, J. The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses 2021, 13, 1115. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Li, Y.H.; Li, J.; Liu, X.E.; Wang, L.; Li, T.; Zhou, Y.H.; Zhou, Y.-H.; Zhuang, H. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: Comparison with results of other viral markers. J. Virol. Methods 2005, 130, 45–50. [Google Scholar] [CrossRef]
- Mu, J.; Xu, J.; Zhang, L.; Shu, T.; Wu, D.; Huang, M.; Ren, Y.; Li, X.; Geng, Q.; Xu, Y.; et al. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci. China Life Sci. 2020, 63, 1413–1416. [Google Scholar] [CrossRef] [Green Version]
- Shah, P.; Canziani, G.A.; Carter, E.P.; Chaiken, I. The Case for S2: The Potential Benefits of the S2 Subunit of the SARS-CoV-2 Spike Protein as an Immunogen in Fighting the COVID-19 Pandemic. Front. Immunol. 2021, 12, 637651. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Legros, V.; Zhou, B.; Siret, E.; Mathieu, C.; Cosset, F.L.; Lavillette, D.; Denolly, S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021, 296, 100111. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachim, A.; Kavian, N.; Cohen, C.A.; Chin, A.W.H.; Chu, D.K.W.; Mok, C.K.P.; Tsang, O.T.Y.; Yeung, Y.C.; Perera, R.A.P.M.; Poon, L.L.M.; et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020, 21, 1293–1301. [Google Scholar] [CrossRef]
- Fu, Y.-Z.; Wang, S.-Y.; Zheng, Z.-Q.; Huang, Y.; Li, W.-W.; Xu, Z.-S.; Wang, Y.-Y. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol. Immunol. 2021, 18, 613–620. [Google Scholar] [CrossRef]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.-D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef]
- Lim, J.M.E.; Hang, S.K.; Hariharaputran, S.; Chia, A.; Tan, N.; Lee, E.S.; Chng, E.; Lim, P.L.; Youmg, B.E.; Lye, D.C.; et al. A comparative characterization of SARS-CoV-2-specific T cells induced by mRNA or inactive virus COVID-19 vaccines. Cell Rep. Med. 2022, 3, 100793. [Google Scholar] [CrossRef]
- Delorey, T.M.; Ziegler, C.G.K.; Heimberg, G.; Normand, R.; Yang, Y.; Segerstolpe, Å.; Abbondanza, D.; Fleming, S.J.; Subremanian, A.; Montoro, D.T.; et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021, 595, 107–113. [Google Scholar] [CrossRef]
- Hou, Y.J.; Okuda, K.; Edwards, C.E.; Martinez, D.R.; Asakura, T.; Dinnon, K.H., 3rd; Kato, T.; Lee, R.E.; Yount, B.L.; Mascenik, T.M.; et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 2020, 182, 429–446.e14. [Google Scholar] [CrossRef] [PubMed]
- Sallenave, J.M.; Guillot, L. Innate Immune Signaling and Proteolytic Pathways in the Resolution or Exacerbation of SARS-CoV-2 in COVID-19: Key Therapeutic Targets? Front. Immunol. 2020, 11, 1229. [Google Scholar] [CrossRef] [PubMed]
- Lowery, S.A.; Sariol, A.; Perlman, S. Innate immune and inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe 2021, 29, 1052–1062. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Sempowski, G.D.; Saunders, K.O.; Acharya, P.; Haynes, B.F. SARS-CoV-2 Neutralizing Antibodies for COVID-19 Prevention and Treatment. Annu. Rev. Med. 2022, 73, 1–16. [Google Scholar] [CrossRef]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef]
- Makarova, O.V.; Diatroptov, M.E.; Serebriakov, S.N.; Malaĭtsev, V.V.; Bogdanova, I.M. Morphofunctional changes of BALB/c and C57BL/6 mice’s immune system under chronic bacterial gram-negative endotoxicosis. Arkh. Patol. 2012, 74, 43–48. [Google Scholar]
- Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Dewitte, H. The dawn of mRNA vaccines: The COVID-19 case. J. Control. Release 2021, 333, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, A.C.; Humbert, M.; Buggert, M. The known unknowns of T cell immunity to COVID-19. Sci. Immunol. 2020, 18, eabe8063. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schafer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Matusali, G.; Colavita, F.; Lapa, D.; Meschi, S.; Bordi, L.; Piselli, P.; Gagliardini, R.; Corpolongo, A.; Nicastri, E.; Antinori, A.; et al. SARS-CoV-2 Serum Neutralization Assay: A Traditional Tool for a Brand-New Virus. Viruses 2021, 13, 655. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Ye, F.; Cheng, M.L.; Feng, Y.; Deng, Y.Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52, 971–977.e3. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Xia, Y.; Liu, X.; Xu, Y.; Lu, P.; Dong, Z.; Liu, J.; Liang, G. A perspective on SARS-CoV-2 virus-like particles vaccines. Int. Immunopharmacol. 2023, 115, 109650. [Google Scholar] [CrossRef]
- Wang, B.; Chen, H.; Jiang, X.; Zhang, M.; Wan, T.; Li, N.; Zhou, X.; Wu, Y.; Yang, F.; Yu, Y.; et al. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood 2004, 104, 200–206. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.Y.; Hwang, K.P.; Kuo, H.K.; Peng, W.J.; Shen, Y.H.; Kuo, B.S.; Huang, J.-H.; Liu, H.; Ho, Y.-H.; Lin, F.; et al. A multitope SARS-CoV-2 vaccine provides long-lasting B cell and T cell immunity against Delta and Omicron variants. J. Clin. Investig. 2022, 132, e157707. [Google Scholar] [CrossRef]
- Le Bert, N.; Clapham, H.E.; Tan, A.T.; Chia, W.N.; Tham, C.Y.L.; Lim, J.M.; Kunasegaran, K.; Tan, L.W.L.; Dutertre, C.-A.; Shankar, N.; et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021, 218, e20202617. [Google Scholar] [CrossRef]
- Li, Z.; Xiang, T.; Liang, B.; Deng, H.; Wang, H.; Feng, X.; Quan, X.; Wang, X.; Li, S.; Lu, S.; et al. Characterization of SARS-CoV-2-Specific Humoral and Cellular Immune Responses Induced by Inactivated COVID-19 Vaccines in a Real-World Setting. Front. Immunol. 2021, 12, 802858. [Google Scholar] [CrossRef]
- Molodtsov, I.A.; Kegeles, E.; Mitin, A.N.; Mityaeva, O.; Musatova, O.E.; Panova, A.E.; Pashenkov, M.V.; Peshkova, I.O.; Alsalloum, A.; Asaad, W.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)-Specific T Cells and Antibodies in Coronavirus Disease 2019 (COVID-19) Protection: A Prospective Study. Clin. Infect Dis. 2022, 75, e1–e9. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, R.; Zeng, F.; Ma, D.; Meng, Z.; Li, X.; Zhang, Z.; Zhang, H.; Li, Q.; Xu, L.; Niu, Z.; et al. Study of the Effects of Several SARS-CoV-2 Structural Proteins on Antiviral Immunity. Vaccines 2023, 11, 524. https://doi.org/10.3390/vaccines11030524
Yue R, Zeng F, Ma D, Meng Z, Li X, Zhang Z, Zhang H, Li Q, Xu L, Niu Z, et al. Study of the Effects of Several SARS-CoV-2 Structural Proteins on Antiviral Immunity. Vaccines. 2023; 11(3):524. https://doi.org/10.3390/vaccines11030524
Chicago/Turabian StyleYue, Rong, Fengyuan Zeng, Danjing Ma, Ziyan Meng, Xinghang Li, Zhenxiao Zhang, Haobo Zhang, Qi Li, Langxi Xu, Zhenye Niu, and et al. 2023. "Study of the Effects of Several SARS-CoV-2 Structural Proteins on Antiviral Immunity" Vaccines 11, no. 3: 524. https://doi.org/10.3390/vaccines11030524





