Early Activation of the Innate Immunity and Specific Cellular Immune Pathways after Vaccination with a Live Intranasal Viral Vaccine and Challenge with Bovine Parainfluenza Type 3 Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Animal Study
2.3. Virus Titration
2.4. Virus Neutralization Serum Antibody Titers
2.5. RT-qPCR Assay
2.6. Statistical Analyses Nasal Shedding of BPI3V and Clinical Scores
2.7. Data Analysis RT-qPCR
3. Results
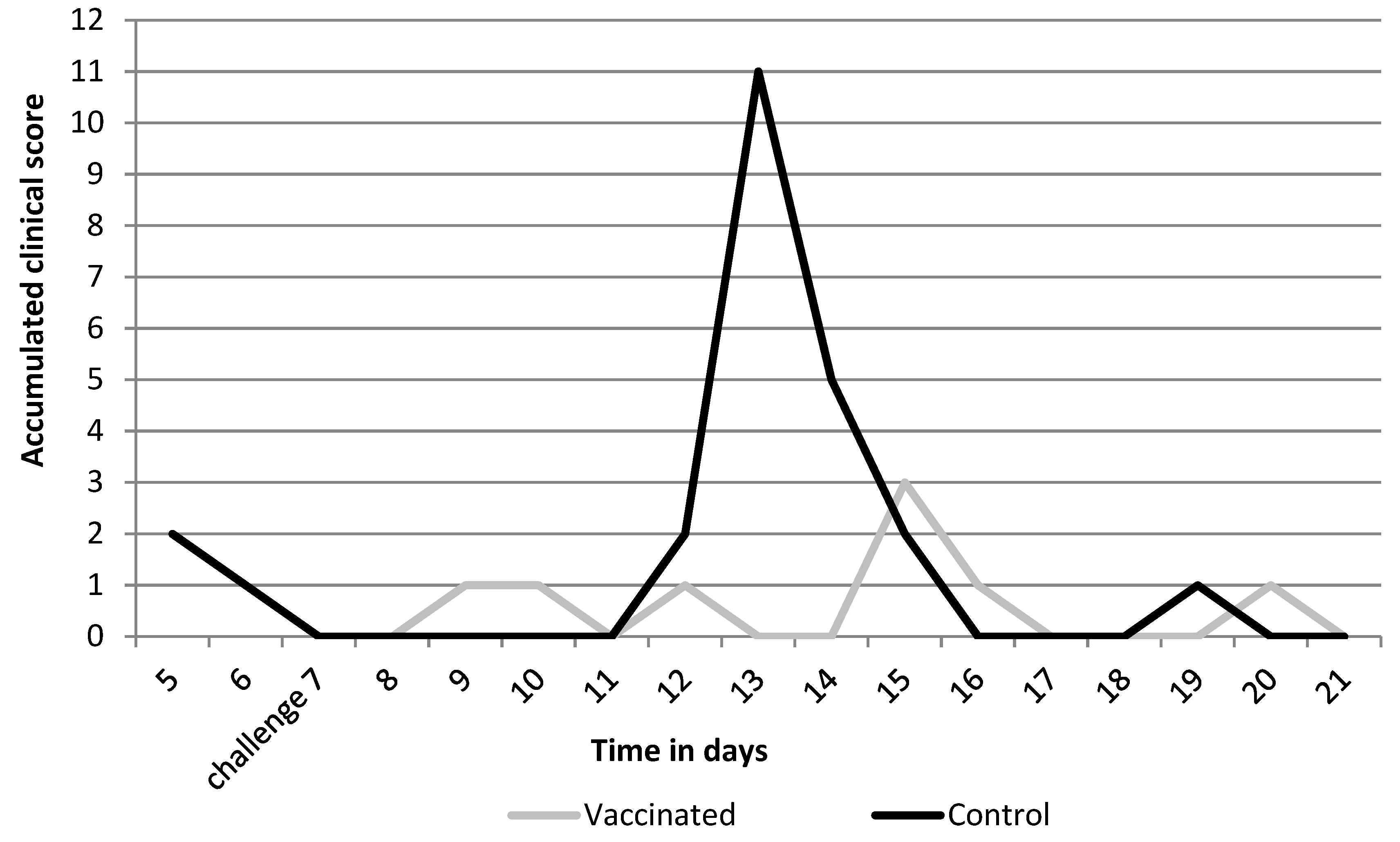
3.1. Efficacy Parameters
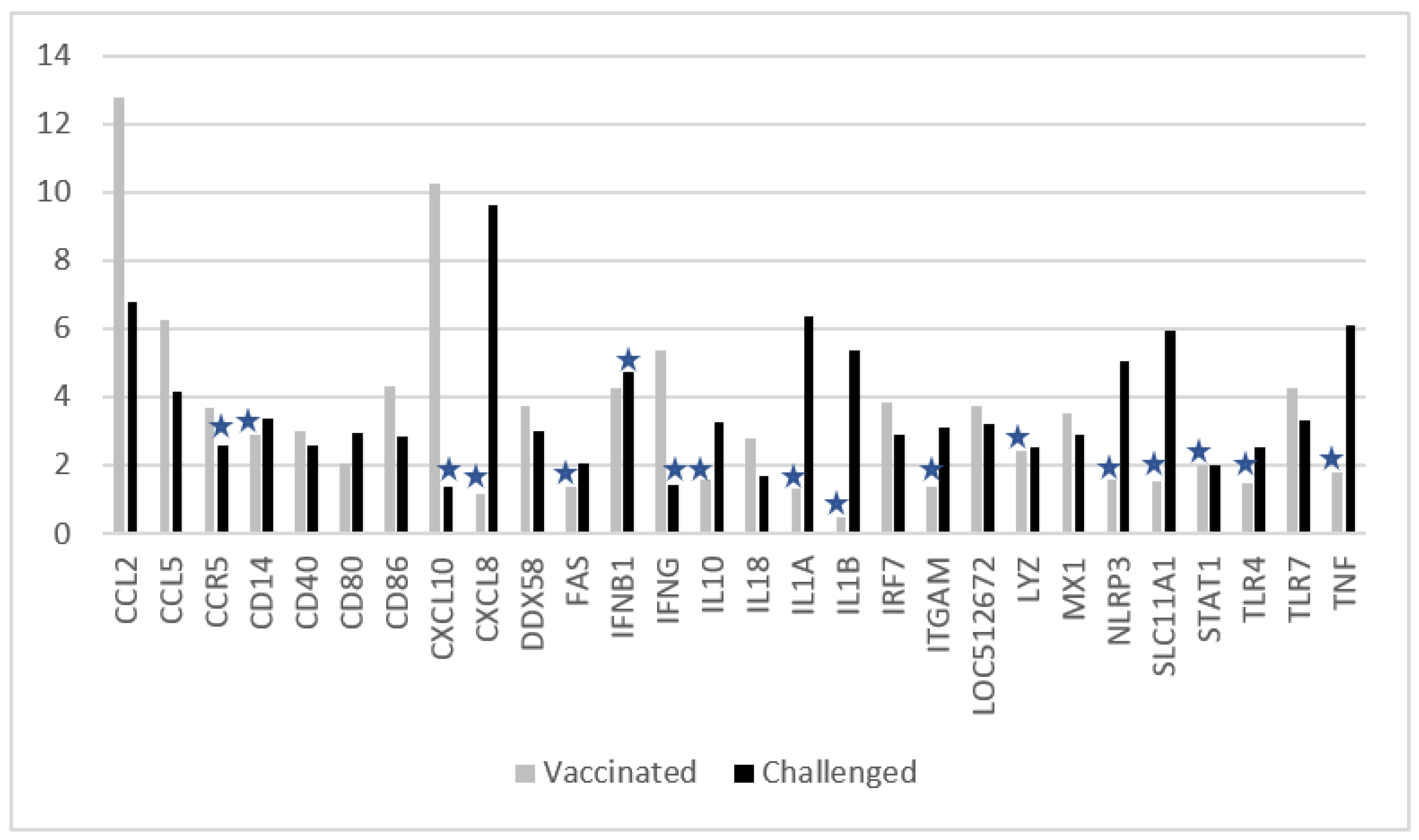
3.2. Changes in Ct Values of Genes after Vaccination, or Challenge, or Vaccination and Challenge
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guerra-Maupome, M.; Palmer, M.V.; McGill, J.L.; Sacco, R.E. Utility of the Neonatal Calf Model for Testing Vaccines and Intervention Strategies for Use against Human RSV Infection. Vaccines 2019, 7, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryson, D.G. Calf Pneumonia. Vet. Clin. N. Am. Food Anim. Pract. 1985, 1, 237–257. [Google Scholar] [CrossRef]
- Fulton, R.W. Bovine respiratory disease research (1983–2009). Anim. Health Res. Rev. 2009, 10, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Bareille, N.; Seegers, H.; Denis, G.; Quillet, J.M.; Assie, S. Impact of respiratory disorders in young bulls during their fattening period on performance and profitability. Renc. Rech. Rumin. 2008, 15, 77–80. [Google Scholar]
- Delabouglise, A.; James, A.; Valarcher, J.-F.; Hagglünd, S.; Raboisson, D.; Rushton, J. Linking disease epidemiology and livestock productivity: The case of bovine respiratory disease in France. PLoS ONE 2017, 12, e0189090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunn, T.R.; Ollivett, T.L.; Renaud, D.L.; Leslie, K.E.; LeBlanc, S.J.; Duffield, T.F.; Kelton, D.F. The effect of lung consolidation, as determined by ultrasonography, on first-lactation milk production in Holstein dairy calves. J. Dairy Sci. 2018, 101, 5404–5410. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, M.R.; Derscheid, R.; Roth, J.A. Innate immunology of bovine respiratory disease. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 215–228. [Google Scholar] [CrossRef] [Green Version]
- Theurer, M.E.; Larson, R.L.; White, B.J. Systematic review and meta-analysis of the effectiveness of commercially available vaccines against bovine herpesvirus, bovine viral diarrhea virus, bovine respiratory syncytial virus, and parainfluenza type 3 virus for mitigation of bovine respiratory disease complex in cattle. J. Am. Vet. Med. Assoc. 2015, 246, 126–142. [Google Scholar]
- Ellis, J.A. The immunology of the bovine respiratory disease complex. Vet. Clin. N. Am. Food Anim. Pract. 2001, 17, 535–550. [Google Scholar] [CrossRef]
- McGill, J.L.; Sacco, R.E. The Immunology of Bovine Respiratory Disease: Recent Advancements Review. Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Nuijten, P.; Cleton, N.; Van der Loop, J.; Makoschey, B.; Vertenten, G. Abstract and poster. In Onset and Duration of Immunity after Vaccination with a New Intranasal BRD Vaccine in Young Calves; European Bovine Congress: ‘s-Hertogenbosch, The Netherlands, 2019. [Google Scholar]
- Gershwin, L.J. Immunology of bovine respiratory syncytial virus infection of cattle. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Guzman, E.; Taylor, G. Immunology of bovine respiratory syncytial virus in calves. Mol. Immunol. 2015, 66, 48–56. [Google Scholar] [CrossRef] [PubMed]
- McGuirk, S.M. Disease management of dairy calves and heifers. Vet. Clin. N. Am. Food Anim. Pract. 2008, 24, 139–153. [Google Scholar] [CrossRef]
- Livak, K.J.; Thomas, D.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real Time Quantitative PCR and the 22DDCT. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Johnston, D.; Earley, B.; McCabe, M.S.; Lemon, K.; Duffy, C.; McMenamy, M.; Cosby, S.L.; Kim, J.; Blackshields, G.; Taylor, J.F.; et al. Experimental challenge with bovine respiratory syncytial virus in dairy calves: Bronchial lymph node transcriptome response. Sci. Rep. 2019, 9, 14736. [Google Scholar] [CrossRef] [PubMed]
- Tizioto, P.C.; Kim, J.; Seabury, C.M.; Schnabel, R.D.; Gershwin, L.J.; Van Eenennaam, A.L.; Toaff-Rosenstein, R.; Neibergs, H.L.; Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team; Taylor, J.F. Immunological Response to Single Pathogen Challenge with Agents of the Bovine Respiratory Disease Complex: An RNA-Sequence Analysis of the Bronchial Lymph Node Transcriptome. PLoS ONE 2015, 10, e0131459. [Google Scholar] [CrossRef]
- Ellis, J.A. Bovine parainfluenza-3 virus. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 575–593. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, M.; Lowe, J.; Aldridge, B. Contribution of the Mucosal Microbiota to Bovine Respiratory Health. Trends Microbiol. 2019, 27, 753–770. [Google Scholar] [CrossRef]
- Wheat, W.; Chow, L.; Rozo, V.; Herman, J.; Still Brooks, K.; Colbath, A.; Hunter, R.; Dowet, S. Non-specific protection from respiratory tract infections in cattle generated by intranasal administration of an innate immune stimulant. PLoS ONE 2020, 15, e0235422. [Google Scholar] [CrossRef]
- Ellis, J.; Gow, S.; West, K.; Waldner, C.; Rhodes, C.; Mutwiri, G.; Rosenberg, H. Response of calves to challenge exposure with virulent bovine respiratory syncytial virus following intranasal administration of vaccines formulated for parenteral administration. J. Am. Vet. Med. Assoc. 2007, 230, 233–243. [Google Scholar] [CrossRef]
- Kimman, T.G.; Westenbrink, F.; Schreuder, B.E.; Straver, P.J. Local and Systemic Antibody Response to Bovine Respiratory Syncytial Virus Infection and Reinfection in Calves with and without Maternal Antibodies. J. Clin. Microbiol. 1987, 25, 1097–1106. [Google Scholar] [CrossRef] [Green Version]
- Rey-Jurado, E.; Bohmwald, K.; Gálvez, N.M.S.; Becerra, D.; Porcelli, S.A.; Carreño, L.J.; Kalergis, A.M. Contribution of NKT cells to the immune response and pathogenesis triggered by respiratory viruses. Virulence 2020, 11, 580–593. [Google Scholar] [CrossRef]
- McGill, J.L.; Sacco, R.E. γδ T cells and the immune response to respiratory syncytial virus infection. Vet. Immunol. Immunopathol. 2016, 181, 24–29. [Google Scholar] [CrossRef] [Green Version]
- Prohl, A.; Wolf, K.; Weber, C.; Müller, K.E.; Menge, C.; Sachse, K.; Rödel, J.; Reinhold, P.; Berndt, A. Kinetics of Local and Systemic Leucocyte and Cytokine Reaction of Calves to Intrabronchial Infection with Chlamydia psittaci. PLoS ONE 2015, 10, e0135161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohlmeier, J.E.; Miller, S.C.; Smith, J.; Lu, B.; Gerard, C.; Cookenham, T.; Roberts, A.D.; Woodland, D.L. CCR5 plays a key role in the early memory CD8+ T cell response to respiratory virus infections. Immunity 2008, 29, 101–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindell, D.M.; Lane, T.E.; Lukacs, N.W. CxCL10/CxCR3-mediated Responses Promote Immunity to Respiratory Syncytial Virus Infection by Augmenting Dendritic Cell and CD8+ T Cell Efficacy. Eur. J. Immunol. 2008, 38, 2168–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberle, K.C.; McGill, J.L.; Reinhardt, T.A.; Sacco, R.E. Parainfluenza Virus 3 Blocks Antiviral Mediators Downstream of the Interferon Lambda Receptor by Modulating Stat1 Phosphorylation. J. Virol. 2015, 90, 2948–2958. [Google Scholar] [CrossRef] [Green Version]
- Valarcher, J.F.; Furze, J.; Wyld, S.; Cook, R.; Conzelmann, K.K.; Taylor, G. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 2003, 77, 8426–8439. [Google Scholar] [CrossRef] [Green Version]


| Title: Log2 VN Titers | Time in Days | ||||
|---|---|---|---|---|---|
| BRSV | BPIV3 | ||||
| Animal | Group | 0 | 0 | 7 | 21 |
| 1 | Vaccinated | 2.0 | 1.0 | 1.0 | 6.0 |
| 2 | Vaccinated | 1.0 | 1.0 | 1.0 | 7.0 |
| 3 | Vaccinated | 1.0 | 1.0 | 1.5 | 7.0 |
| 4 | Vaccinated | 1.0 | 1.0 | 1.0 | 9.0 |
| 5 | Vaccinated | 1.5 | 1.5 | 1.5 | 7.0 |
| 6 | Vaccinated | 1.0 | 1.0 | 2.0 | 7.0 |
| Average | 1.3 | 1.1 | 1.3 | 7.3 | |
| 7 | Controls | 1.0 | 1.0 | 1.0 | 7.0 |
| 8 | Controls | 1.0 | 1.0 | 1.0 | 3.5 |
| 9 | Controls | 1.0 | 1.0 | 1.0 | 5.5 |
| 10 | Controls | 1.0 | 1.0 | 1.0 | 6.0 |
| 11 | Controls | 1.0 | 1.0 | 1.0 | 3.0 |
| 12 | Controls | 1.0 | 1.0 | 1.0 | 4.5 |
| 13 | Controls | 1.0 | 1.0 | 1.0 | 5.0 |
| Average | 1.0 | 1.0 | 1.0 | 4.9 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuijten, P.; Cleton, N.; van der Loop, J.; Makoschey, B.; Pulskens, W.; Vertenten, G. Early Activation of the Innate Immunity and Specific Cellular Immune Pathways after Vaccination with a Live Intranasal Viral Vaccine and Challenge with Bovine Parainfluenza Type 3 Virus. Vaccines 2022, 10, 104. https://doi.org/10.3390/vaccines10010104
Nuijten P, Cleton N, van der Loop J, Makoschey B, Pulskens W, Vertenten G. Early Activation of the Innate Immunity and Specific Cellular Immune Pathways after Vaccination with a Live Intranasal Viral Vaccine and Challenge with Bovine Parainfluenza Type 3 Virus. Vaccines. 2022; 10(1):104. https://doi.org/10.3390/vaccines10010104
Chicago/Turabian StyleNuijten, Piet, Natalie Cleton, Jeroen van der Loop, Birgit Makoschey, Wilco Pulskens, and Geert Vertenten. 2022. "Early Activation of the Innate Immunity and Specific Cellular Immune Pathways after Vaccination with a Live Intranasal Viral Vaccine and Challenge with Bovine Parainfluenza Type 3 Virus" Vaccines 10, no. 1: 104. https://doi.org/10.3390/vaccines10010104






