Detection of Proteins Using Nano Magnetic Particle Accumulation-Based Signal Amplification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Procedures
2.1.1. Immobilization of BBSA on Gold Chips
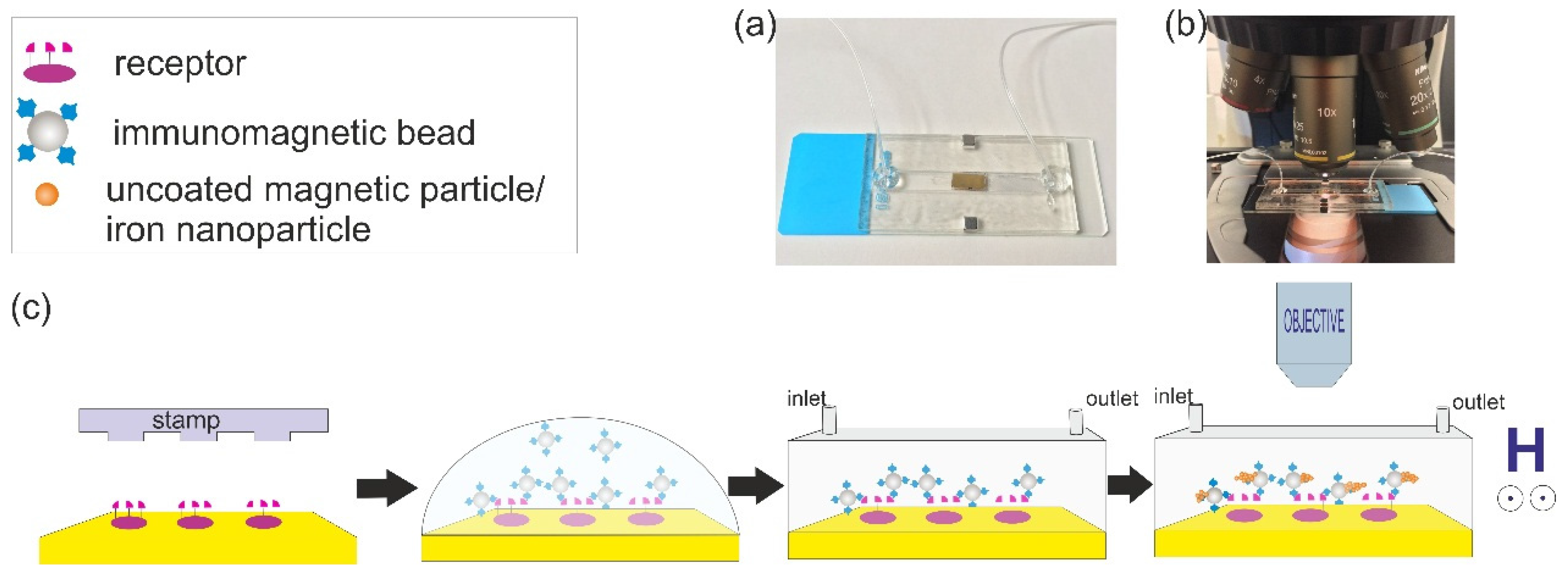
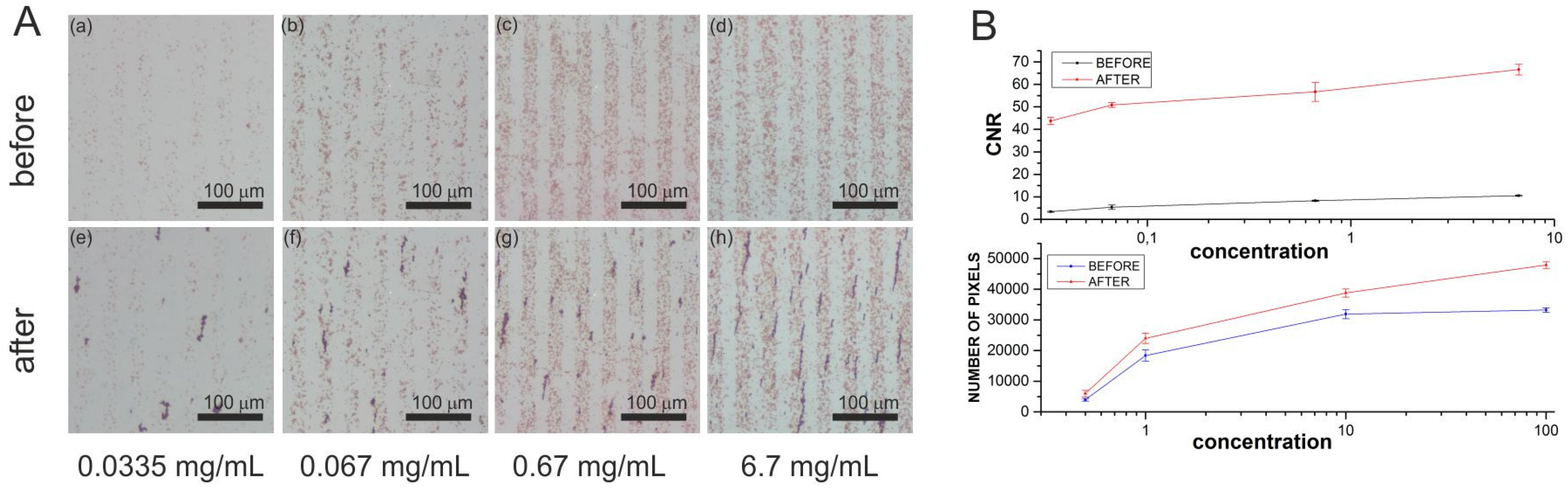
2.1.2. First Set of Experiments: Magnetic Accumulation of Iron Nano Particles around the Immobilized Streptavidin-Coated Magnetic Beads
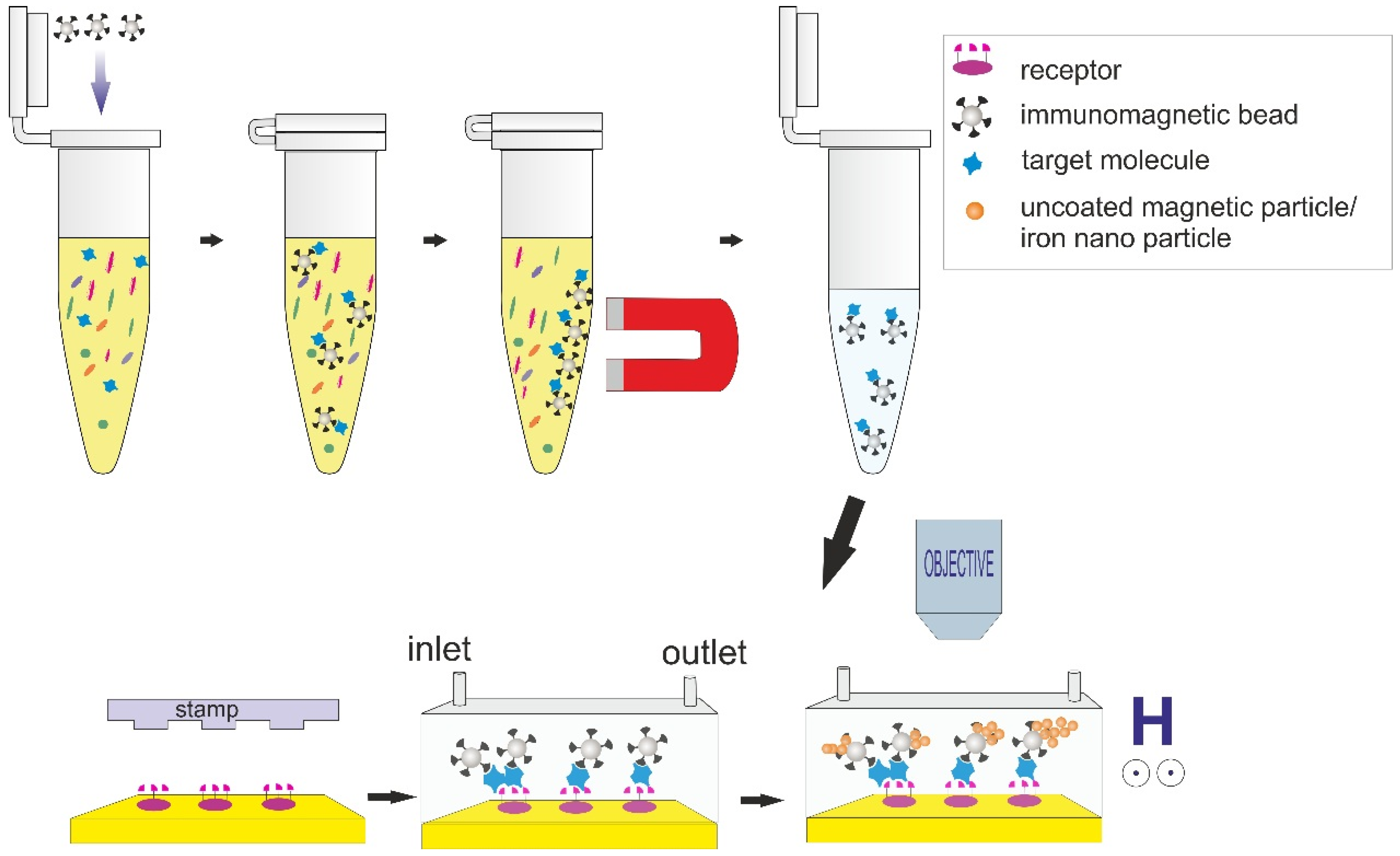
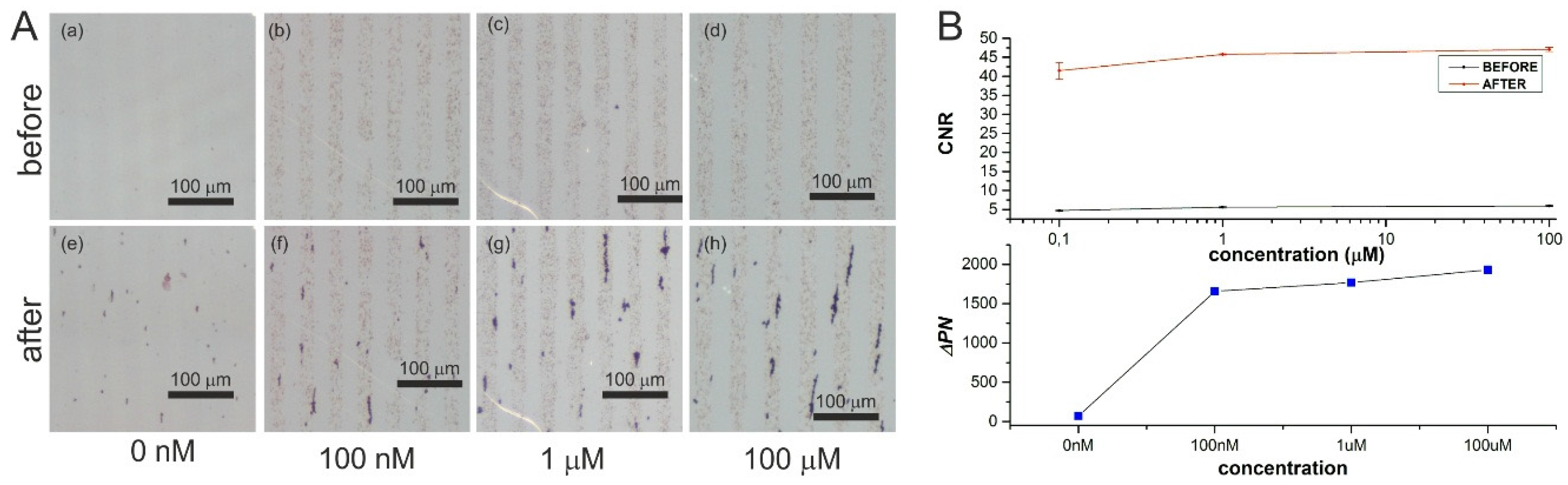
2.1.3. Second Set of Experiments: Detection of Streptavidin (Target Capture, Separation, Sandwich Assay Formation, and then Magnetic Signal Amplification)
2.1.4. Quantification Methods
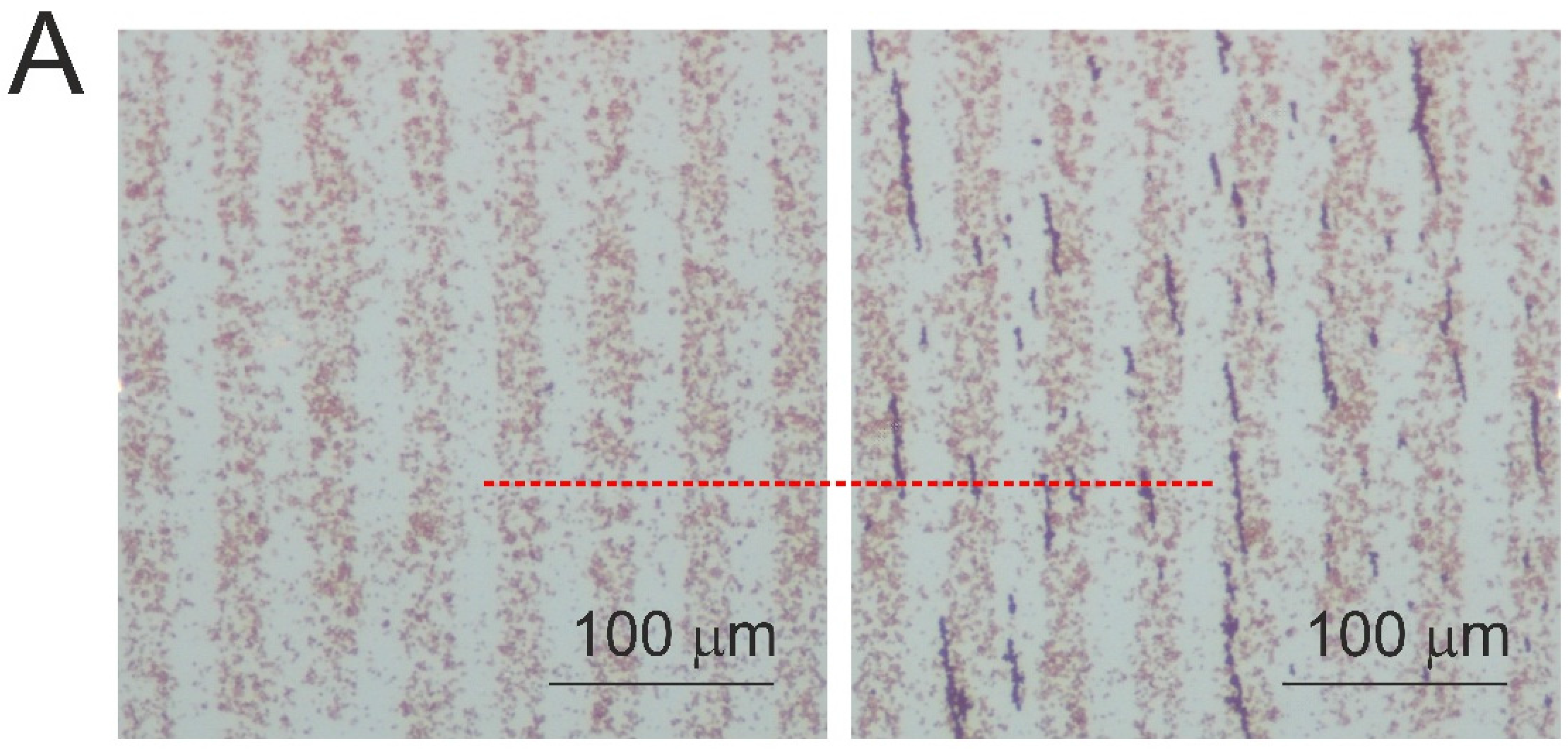
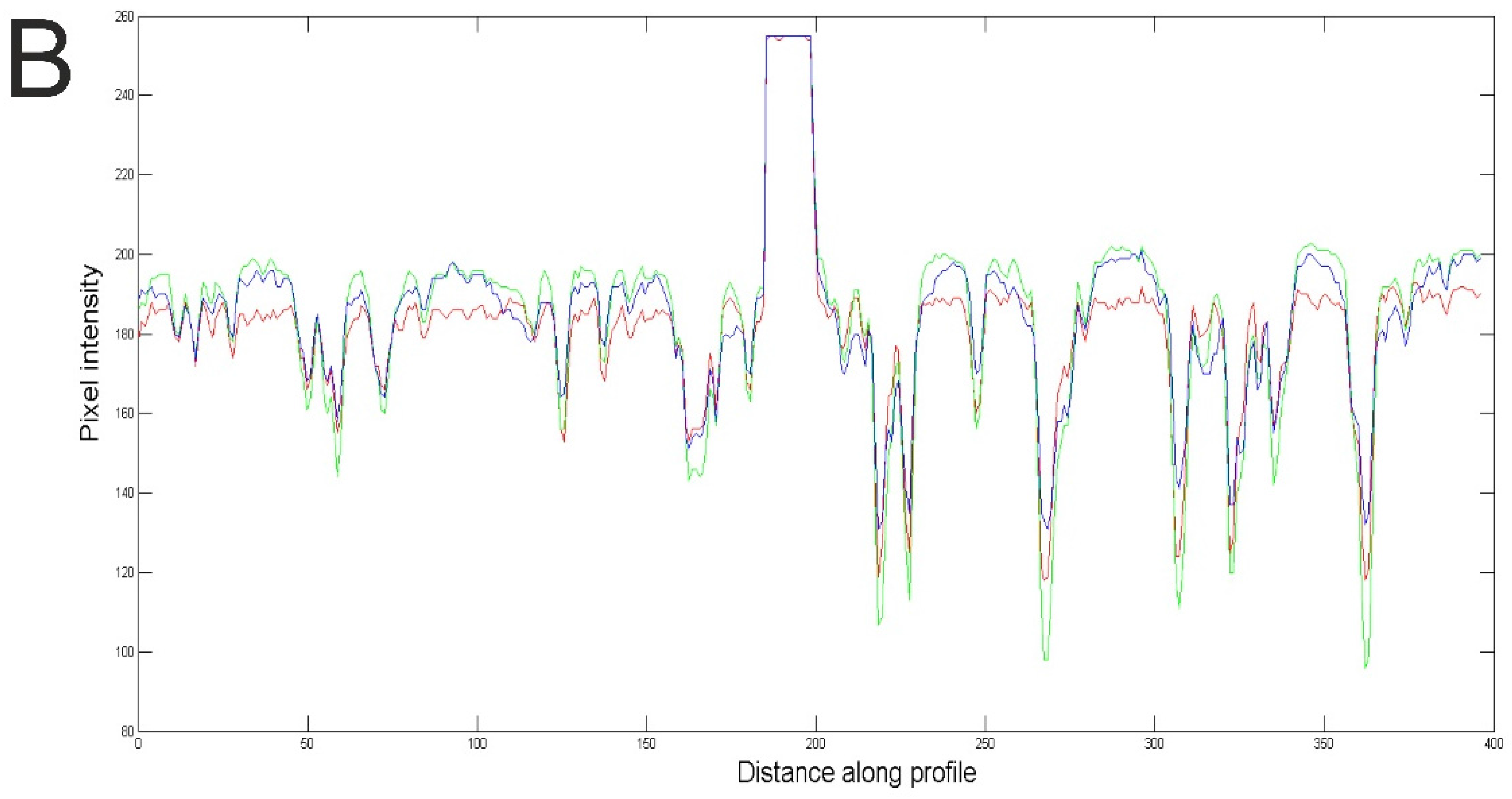
3. Results
4. Discussion and Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- İçöz, K.; Savran, C. Nanomechanical biosensing with immunomagnetic separation. Appl. Phys. Lett. 2010, 97. [Google Scholar] [CrossRef]
- Icoz, K.; Iverson, B.D.; Savran, C. Noise analysis and sensitivity enhancement in immunomagnetic nanomechanical biosensors. Appl. Phys. Lett. 2008, 93. [Google Scholar] [CrossRef]
- Janssen, X.J.A.; van IJzendoorn, L.J.; Prins, M.W.J. On-chip manipulation and detection of magnetic particles for functional biosensors. Biosens. Bioelectron. 2008, 23, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Inglis, D.W.; Riehn, R.; Austin, R.H.; Sturm, J.C. Continuous microfluidic immunomagnetic cell separation. Appl. Phys. Lett. 2004, 85, 5093–5095. [Google Scholar] [CrossRef]
- Earhart, C.M.; Hughes, C.E.; Gaster, R.S.; Ooi, C.C.; Wilson, R.J.; Zhou, L.Y.; Humke, E.W.; Xu, L.; Wong, D.J.; Willingham, S.B.; et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip 2014, 14, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Jalal, S.I.; Huang, W.; Mahmood, A.; Matei, D.E.; Savran, C.A. High-throughput immunomagnetic cell detection using a microaperture chip system. 2014, 14, 3008–3013. [Google Scholar] [CrossRef]
- Zborowski, M.; Chalmers, J.J. Rare cell separation and analysis by magnetic sorting. Anal. Chem. 2011, 83, 8050–8056. [Google Scholar] [CrossRef] [PubMed]
- Forbes, T.P.; Forry, S.P. Microfluidic magnetophoretic separations of immunomagnetically labeled rare mammalian cells. Lab Chip 2012, 12, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Nath, P.; Moore, L.R.; Zborowski, M.; Roy, S.; Fleischman, A. A method to obtain uniform magnetic-field energy density gradient distribution using discrete pole pieces for a microelectromechanical-system- based magnetic cell separator. J. Appl. Phys. 2006, 99, 2004–2007. [Google Scholar] [CrossRef]
- Choi, J.-W.; Oh, K.W.; Thomas, J.H.; Heineman, W.R.; Halsall, H.B.; Nevin, J.H.; Helmicki, A.J.; Henderson, H.T.; Ahn, C.H. An integrated microfluidic biochemical detection system for protein analysis with magnetic bead-based sampling capabilities. Lab Chip 2002, 2, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Ruangchaithaweesuk, S.; Yao, L.; Xu, S. Detecting molecules and cells labeled with magnetic particles using an atomic magnetometer. J. Nanopart. Res. 2012, 14. [Google Scholar] [CrossRef]
- Hao, L.; Aßmann, C.; Gallop, J.C.; Cox, D.; Ruede, F.; Kazakova, O.; Josephs-Franks, P.; Drung, D.; Schurig, T. Detection of single magnetic nanobead with a nano-superconducting quantum interference device. Appl. Phys. Lett. 2011, 98. [Google Scholar] [CrossRef] [Green Version]
- Rife, J.C.; Miller, M.M.; Sheehan, P.E.; Tamanaha, C.R.; Tondra, M.; Whitman, L.J. Design and performance of GMR sensors for the detection of magnetic microbeads in biosensors. Sens. Actuators A Phys. 2003, 107, 209–218. [Google Scholar] [CrossRef]
- Huang, H.-T.; Ger, T.-R.; Lin, Y.-H.; Wei, Z.-H. Single cell detection using a magnetic zigzag nanowire biosensor. Lab Chip 2013, 13, 3098–3104. [Google Scholar] [CrossRef] [PubMed]
- Besse, P.-A.; Boero, G.; Demierre, M.; Pott, V.; Popovic, R. Detection of a single magnetic microbead using a miniaturized silicon Hall sensor. Appl. Phys. Lett. 2002, 80. [Google Scholar] [CrossRef]
- Lee, J.; Icoz, K.; Roberts, A.; Ellington, A.D.; Savran, C.A. Diffractometric detection of proteins using microbead-based rolling circle amplification. Anal. Chem. 2010, 82, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.M.; Leslie, D.C.; Domansky, K.; Jain, A.; Yung, C.; Cho, M.; Workman, S.; Super, M.; Ingber, D.E. A microdevice for rapid optical detection of magnetically captured rare blood pathogens. Lab Chip 2014, 14, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Mai, T.D.; Pereiro, I.; Hiraoui, M.; Viovy, J.-L.; Descroix, S.; Taverna, M.; Smadja, C. Magneto-immunocapture with on-bead fluorescent labeling of amyloid-β peptides: Towards a microfluidized-bed-based operation. Analyst 2015, 140, 5891–5900. [Google Scholar] [CrossRef] [PubMed]
- Fan, A.; Lau, C.; Lu, J. Magnetic bead-based chemiluminescent metal immunoassay with a colloidal gold label. Anal. Chem. 2005, 77, 3238–3242. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, R.; Patel, V.; Chikkaveeraiah, B.V.; Munge, B.S.; Cheong, S.C.; Zain, R.B.; Abraham, M.T.; Dey, D.K.; Gutkind, J.S.; Rusling, J.F. Ultrasensitive detection of cancer biomarkers in the clinic by use of a nanostructured microfluidic array. Anal. Chem. 2012, 84, 6249–6255. [Google Scholar] [CrossRef] [PubMed]
- Mzava, O.; Taş, Z.; İçöz, K. Magnetic micro/nanoparticle flocculation-based signal amplification for biosensing. Int. J. Nanomed. 2016, 11, 2619–2631. [Google Scholar]
- Acharya, G.; Chang, C.L.; Doorneweerd, D.D.; Vlashi, E.; Henne, W.A.; Hartmann, L.C.; Low, P.S.; Savran, C.A. Immunomagnetic diffractometry for detection of diagnostic serum markers. J. Am. Chem. Soc. 2007, 129, 15824–15829. [Google Scholar] [CrossRef] [PubMed]
- ImageJ. Available online: https://imagej.nih.gov/ij/ (accessed on 23 September 2016).
- Gomi, T. Dual-energy subtraction X-ray digital tomosynthesis: Basic physical evaluation. Open J. Med. Imaging 2012, 2, 111–117. [Google Scholar] [CrossRef]
- Acharya, G.; Chang, C.L.; Holland, D.P.; Thompson, D.H.; Savran, C.A. Rapid detection of S-adenosyl homocysteine using self-assembled optical diffraction gratings. Angew. Chem. Int. Ed. 2008, 47, 1051–1053. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.L.; Acharya, G.; Savran, C.A. In situ assembled diffraction grating for biomolecular detection. Appl. Phys. Lett. 2007, 90. [Google Scholar] [CrossRef]
- Arpa, A.; Wetzstein, G.; Lanman, D.; Raskar, R. Single lens off-chip cellphone microscopy. In IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops; IEEE: Piscataway, NJ, USA, 2012; pp. 23–28. [Google Scholar]
- Seo, S.; Su, T.-W.; Tseng, D.K.; Erlinger, A.; Ozcan, A. Lensfree holographic imaging for on-chip cytometry and diagnostics. Lab Chip 2009, 9, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.J.; Chu, K.; Espenson, A.R.; Rahimzadeh, M.; Gryshuk, A.; Molinaro, M.; Dwyre, D.M.; Lane, S.; Matthews, D.; Wachsmann-Hogiu, S. Cell-phone-based platform for biomedical device development and education applications. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- İçöz, K. Image processing and cell phone microscopy to analyze the immunomagnetic beads on micro-contact printed gratings. Appl. Sci. 2016, 6. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
İçöz, K.; Mzava, O. Detection of Proteins Using Nano Magnetic Particle Accumulation-Based Signal Amplification. Appl. Sci. 2016, 6, 394. https://doi.org/10.3390/app6120394
İçöz K, Mzava O. Detection of Proteins Using Nano Magnetic Particle Accumulation-Based Signal Amplification. Applied Sciences. 2016; 6(12):394. https://doi.org/10.3390/app6120394
Chicago/Turabian Styleİçöz, Kutay, and Omary Mzava. 2016. "Detection of Proteins Using Nano Magnetic Particle Accumulation-Based Signal Amplification" Applied Sciences 6, no. 12: 394. https://doi.org/10.3390/app6120394
APA Styleİçöz, K., & Mzava, O. (2016). Detection of Proteins Using Nano Magnetic Particle Accumulation-Based Signal Amplification. Applied Sciences, 6(12), 394. https://doi.org/10.3390/app6120394





