The Role of the TLR4-MyD88 Signaling Pathway in the Immune Response of the Selected Scallop Strain “Hongmo No. 1” to Heat Stress
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Information
2.2. Heat Stress Treatment
2.3. Injection of Lipopolysaccharide (LPS) at 24 °C and 32 °C
2.4. RNA Extraction
2.5. Molecular Cloning and Sequencing
2.6. Bioinformatic Analysis
2.7. Real-Time Quantitative PCR (RT-PCR)
2.8. RNA Interference (RNAi) of HMTLR4 Gene Expression In Vivo
2.9. Enzyme-Linked Immunosorbent Assay (ELISA)
3. Results
3.1. Gene Cloning and Sequence Analysis of HMTLR4, HMMyD88 and HMTRAF6
3.2. Phylogenetic Analysis of HMTLR4, HMMyD88 and HMTRAF6
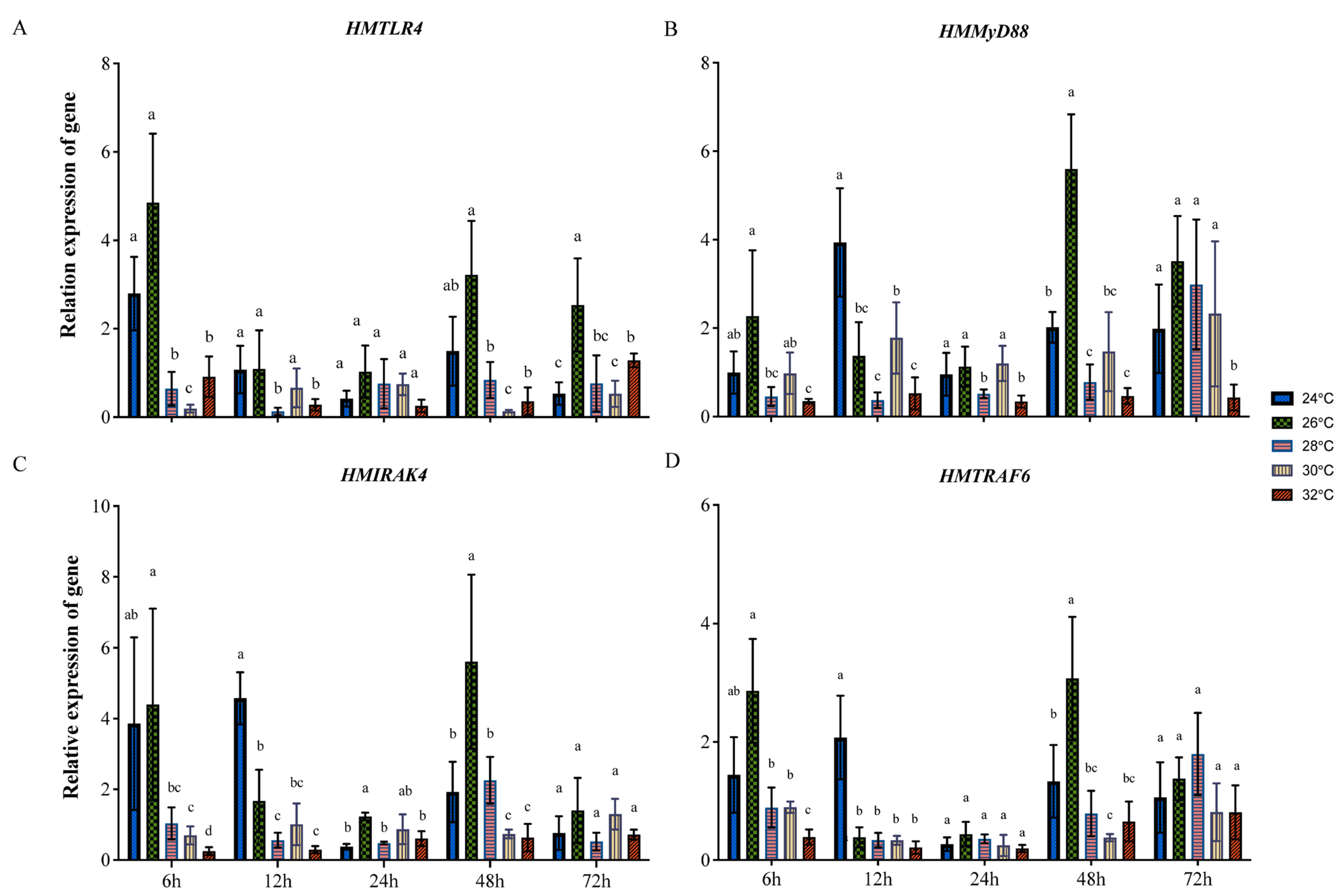
3.3. Expression of HMTLR4, HMMyD88, HMIRAK4 and HMTRAF6 at Different Temperatures
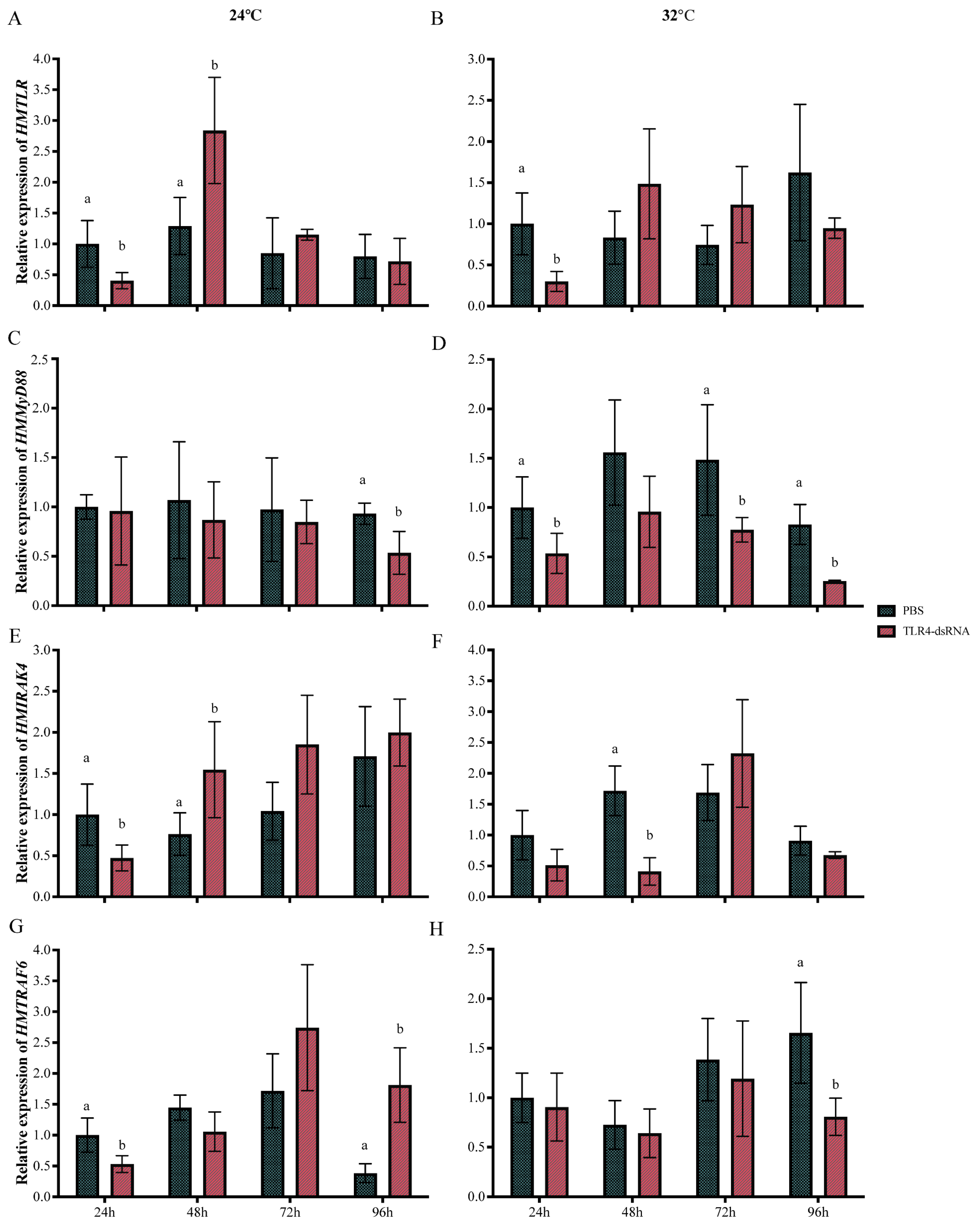
3.4. The Expression of Genes in the TLR4-MyD88 Pathway after LPS Injection at 24 °C and 32 °C
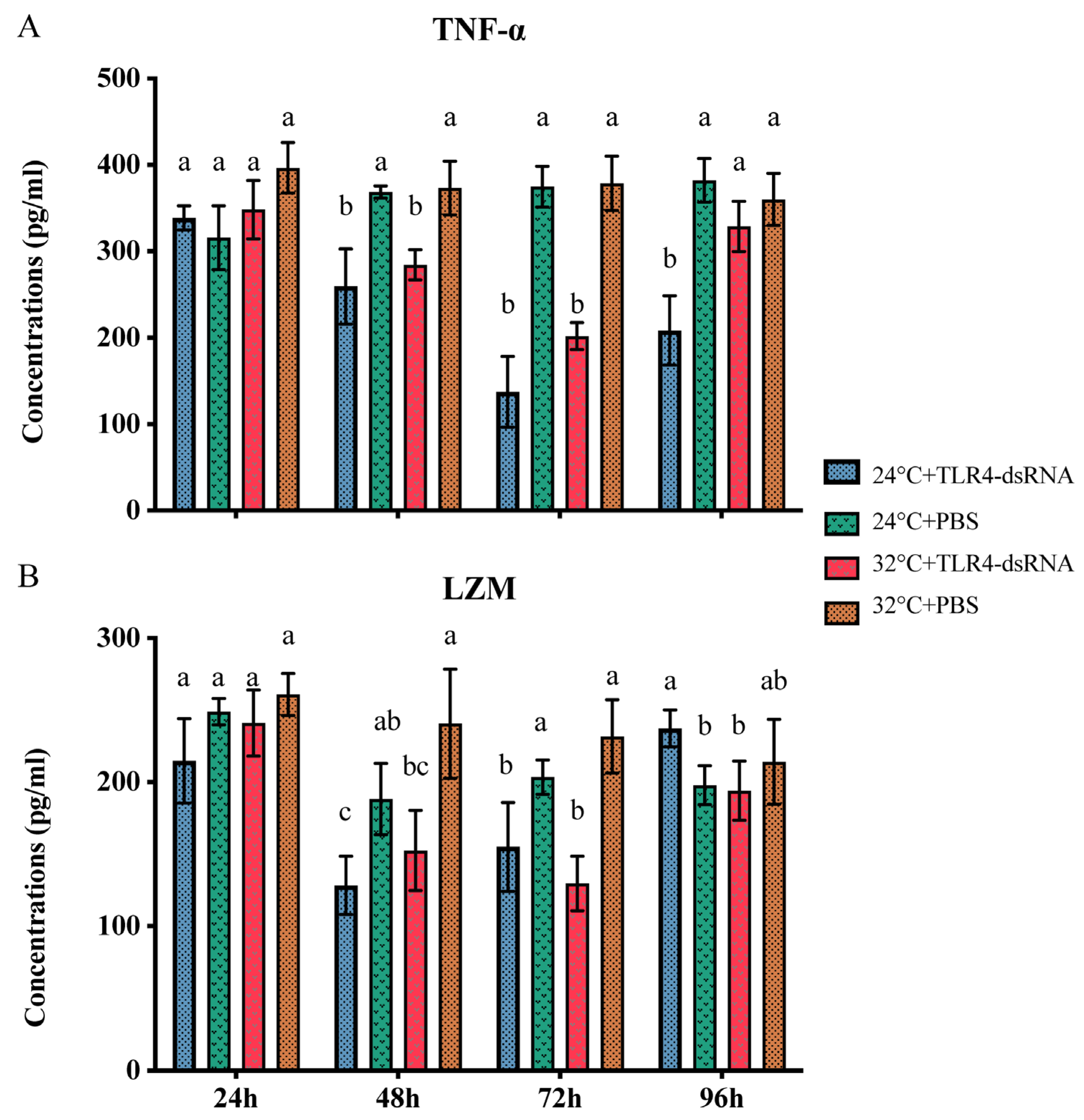
3.5. Levels of TNF-α and LZM under Different Stress Conditions
3.6. The Effect of HMTLR4 Suppression on the Expression of Downstream Genes and the Levels of TNF-α and LZM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Perkins-Kirkpatrick, S.E.; Stone, D.A.; Mitchell, D.M.; Rosier, S.; King, A.D.; Lo, Y.T.E.; Pastor-Paz, J.; Frame, D.; Wehner, M. On the attribution of the impacts of extreme weather events to anthropogenic climate change. Environ. Res. Lett. 2022, 17, 024009. [Google Scholar] [CrossRef]
- Islam, M.J.; Kunzmann, A.; Slater, M.J. Responses of aquaculture fish to climate change-induced extreme temperatures: A review. J. World Aquac. Soc. 2022, 53, 314–366. [Google Scholar] [CrossRef]
- Dégremont, L.; Bédier, E.; Soletchnik, P.; Ropert, M.; Huvet, A.; Moal, J.; Samain, J.-F.; Boudry, P. Relative importance of family, site, and field placement timing on survival, growth, and yield of hatchery-produced Pacific oyster spat (Crassostrea gigas). Aquaculture 2005, 249, 213–229. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, X.; Hu, X.; Gong, J.; Hwang, H.; Mai, K. Effects of temperature on non-specific immune parameters in two scallop species: Argopecten irradians (Lamarck 1819) and Chlamys farreri (Jones & Preston 1904). Aquac. Res. 2004, 35, 678–682. [Google Scholar] [CrossRef]
- Ferreira-Rodríguez, N.; Fernández, I.; Cancela, M.L.; Pardo, I. Multibiomarker response shows how native and non-native freshwater bivalves differentially cope with heat-wave events. Aquat. Conserv. 2018, 28, 934–943. [Google Scholar] [CrossRef]
- Wang, C.; Liu, B.; Li, J.; Liu, S.; Li, J.; Hu, L.; Fan, X.; Du, H.; Fang, H. Introduction of the Peruvian scallop and its hybridization with the bay scallop in China. Aquaculture 2011, 310, 380–387. [Google Scholar] [CrossRef]
- Wang, C.; Liu, B.; Liu, X.; Ma, B.; Zhao, Y.; Zhao, X.; Liu, F.; Liu, G. Selection of a new scallop strain, the Bohai Red, from the hybrid between the bay scallop and the Peruvian scallop. Aquaculture 2017, 479, 250–255. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Wang, C.; Yao, G.; Zhang, K.; Zhan, J.; Lu, W.; Zhong, M.; Liufu, S.; Fang, J. Growth performance and model fitting of the selected strain of scallop “Hongmo No. 1” cultivated during different seasons. Aquaculture 2023, 578, 740104. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C.A. Innate immunity: The virtues of a nonclonal system of recognition. Cell 1997, 91, 295–298. [Google Scholar] [CrossRef]
- Hoshino, K.; Takeuchi, O.; Kawai, T.; Sanjo, H.; Ogawa, T.; Takeda, Y.; Takeda, K.; Akira, S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the Lps gene product. J. Immunol. 1999, 162, 3749–3752. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signaling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Saco, A.; Novoa, B.; Greco, S.; Gerdol, M.; Figueras, A. Bivalves present the largest and most diversified repertoire of Toll-like receptors in the animal kingdom, suggesting broad-spectrum pathogen recognition in marine waters. Mol. Biol. Evol. 2023, 40, msad133. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.C.; Young, D.W.; Golenbock, D.T.; Christ, W.J.; Gusovsky, F. Toll-like receptor-4 mediates lipopolysaccharide- induced signal transduction. J. Biol. Chem. 1999, 274, 10689–10692. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Preston-Hurlburt, P.; Kopp, E.; Stadlen, A.; Chen, C.; Ghosh, S.; Janeway, C.A., Jr. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 1998, 2, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Muzio, M.; Natoli, G.; Saccani, S.; Levrero, M.; Mantovani, A. The human Toll signaling pathway: Divergence of nuclear factor κB and JNK/SAPK activation upstream of tumor necrosis factor receptor–associated factor 6 (TRAF6). J. Exp. Med. 1998, 187, 2097–2101. [Google Scholar] [CrossRef]
- Horng, T.; Barton, G.; Medzhitov, R. TIRAP: An adapter molecule in the Toll signaling pathway. Nat. Immunol. 2001, 2, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Venier, P.; Edomi, P. Diversity and evolution of TIR-domain-containing proteins in bivalves and Metazoa: New insights from comparative genomics. Dev. Comp. Immunol. 2017, 70, 145–164. [Google Scholar] [CrossRef]
- Xing, Q.; Liao, H.; Xun, X.; Wang, J.; Zhang, Z.; Yang, Z.; Huang, X.; Bao, Z. Genome-wide identification, characterization and expression analyses of TLRs in Yesso scallop (Patinopecten yessoensis) provide insight into the disparity of responses to acidifying exposure in bivalves. Fish Shellfish Immun. 2017, 68, 280–288. [Google Scholar] [CrossRef]
- Chen, J.; Lin, J.; Yu, F.; Zhong, Z.; Liang, Q.; Pang, H.; Wu, S. Transcriptome analysis reveals the function of TLR4-MyD88 pathway in immune response of Crassostrea hongkongensis against Vibrio Parahemolyticus. Aquacult. Rep. 2022, 25, 101253. [Google Scholar] [CrossRef]
- Yu, F.; Chen, J.; Lin, J.; Zhong, Z.; Lu, Y.; Zeng, X.; Lei, X. TLR4 involved in immune response against Vibrio Parahaemolyticus by MyD88-dependent pathway in Crassostrea hongkongensis. Fish Shellfish Immun. 2023, 134, 108591. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, M.; Liang, H.; Shen, C.; Zhang, B.; Liang, B. Comparative proteomics and transcriptomics illustrate the allograft-induced stress response in the pearl oyster (Pinctada fucata martensii). Fish Shellfish Immun. 2022, 121, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Li, Q.; Yu, H. Integrated analysis of miRNA and mRNA expression profiles identifies potential regulatory interactions during sexual development of Pacific oyster Crassostrea gigas. Aquaculture 2022, 546, 737294. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, N.; Tanaka, T.; Enkhbayar, P.; Mikami, T.; Taga, M.; Yamada, K.; Kuroki, Y. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genom. 2007, 8, 124. [Google Scholar] [CrossRef]
- Matsushima, N.; Miyashita, H.; Enkhbayar, P.; Kretsinger, R.H. Comparative geometrical analysis of leucine-rich repeat structures in the nod-like and toll-like receptors in vertebrate innate immunity. Biomolecules 2015, 5, 1955–1978. [Google Scholar] [CrossRef]
- Hughes, A.L.; Piontkivska, H. Functional diversification of the toll-like receptor gene family. Immunogenetics 2008, 60, 249–256. [Google Scholar] [CrossRef]
- Zhang, Q.; Zmasek, C.M.; Godzik, A. Domain architecture evolution of pattern recognition receptors. Immunogenetics 2010, 62, 263–272. [Google Scholar] [CrossRef]
- Buckley, K.M.; Rast, J.P. Dynamic evolution of toll-like receptor multigene families in echinoderms. Front. Immunol. 2012, 3, 136. [Google Scholar] [CrossRef]
- Wesche, H.; Henzel, W.J.; Shillinglaw, W.; Li, S.; Cao, Z. MyD88: An adapter that recruits IRAK to the IL-1 receptor complex. Immunity 1997, 7, 837–847. [Google Scholar] [CrossRef]
- Zhang, Y.; He, X.; Yu, F.; Xiang, Z.; Li, J.; Thorpe, K.L.; Yu, Z. Characteristic and functional analysis of toll-like receptors (TLRs) in the lophotrocozoan, Crassostrea gigas, reveals ancient origin of TLR-mediated innate immunity. PLoS ONE 2013, 8, e76464. [Google Scholar] [CrossRef]
- Ju, X.-H.; Xu, H.-J.; Yong, Y.-H.; An, L.-L.; Jiao, P.-R.; Liao, M. Heat stress upregulation of toll-like receptors 2/4 and acute inflammatory cytokines in peripheral blood mononuclear cell (PBMC) of Bama miniature pigs: An in vivo and in vitro study. Animal 2014, 8, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Binder, R.J.; Suto, R.; Anderson, K.M.; Srivastava, P.K. Necrotic but not apoptotic cell death releases heat shock proteins, which deliver a partial maturation signal to dendritic cells and activate the NF-κB pathway. Int. Immunol. 2000, 12, 1539–1546. [Google Scholar] [CrossRef] [PubMed]
- Martine, P.; Rébé, C. Heat Shock Proteins and Inflammasomes. Int. J. Mol. Sci. 2019, 20, 4508. [Google Scholar] [CrossRef] [PubMed]
- Cantet, J.M.; Yu, Z.; Ríus, A.G. Heat Stress-Mediated Activation of Immune–Inflammatory Pathways. Antibiotics 2021, 10, 1285. [Google Scholar] [CrossRef] [PubMed]
- Basu, M.; Paichha, M.; Swain, B.; Lenka, S.S.; Singh, S.; Chakrabarti, R.; Samanta, M. Modulation of TLR2, TLR4, TLR5, NOD1 and NOD2 receptor gene expressions and their downstream signaling molecules following thermal stress in the Indian major carp catla (Catla catla). Biotech 2015, 5, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Ciesielska, A.; Matyjek, M.; Kwiatkowska, K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 2021, 78, 1233–1261. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Sinha, D.; Mukherjee, S.; Biswas, R.; Biswas, T. LPS stimulates and Hsp70 down-regulates TLR4 to orchestrate differential cytokine response of culture-differentiated innate memory CD8+ T cells. Cytokine 2015, 73, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yang, J.; Zhou, Z. A primitive Toll-like receptor signaling pathway in mollusk Zhikong scallop Chlamys farreri. Dev. Comp. Immunol. 2011, 35, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Parisi, M.; Baranzini, N.; Dara, M.; La Corte, C.; Vizioli, J.; Cammarata, M. AIF-1 and RNASET2 are involved in the inflammatory response in the Mediterranean mussel Mytilus galloprovincialis following Vibrio infection. Fish Shellfish Immunol. 2022, 127, 109–118. [Google Scholar] [CrossRef] [PubMed]


| Primer | Sequence (5′–3′) | Application |
|---|---|---|
| HMTLR4-5-1 | TGGAGACAGAGCCGTAACCCACG | 5′RACE |
| HMTLR4-5-2 | GGTGGAGACAGAGCCGTAACCCA | 5′RACE |
| HMMyD88-5-1 | AATTGTGGACACTATAGTTATGCTTGAC | 5′RACE |
| HMMyD88-5-2 | CATCACCGCCTTTAACCACAAT | 5′RACE |
| HMTRAF6-5-1 | GCCATTGCCTGTCTTTCAACCGT | 5′RACE |
| HMTLR4-3-1 | TAGATTTGTCGGGTGAAGTCGGTAGC | 3′RACE |
| HMTLR4-3-2 | TTCACGCTGGGAAGGTATCTGCT | 3′RACE |
| HMMyD88-3-1 | TTACAGGGCAAGTTGGGGTGT | 3′RACE |
| HMMyD88-3-2 | AAATTATGATGATTTCGCAGCTATG | 3′RACE |
| HMTRAF6-3-1 | GAAGGCTGTGATACGCAGGTGGT | 3′RACE |
| HMTLR4-F | CCTTACCTGTATTTCCCCCC | RT-PCR |
| HMTLR4-R | GGTTTCTGACTGGACCCTTC | RT-PCR |
| HMMyD88-F | TTTTAGGAACAGGCTTTA | RT-PCR |
| HMMyD88-R | TGTGGACACGTTGATAAT | RT-PCR |
| HMIRAK4-F | TGTCGTACTGCTGCCTGAT | RT-PCR |
| HMIRAK4-R | TGGAAGCGTAGACTGGAGA | RT-PCR |
| HMTRAF6-F | TCAGGTGAAGCGATTGGG | RT-PCR |
| HMTRAF6-R | TCCTGGTGGAAAGGTAAT | RT-PCR |
| β-actin-F | CCGTGACTTGACCGATTACC | RT-PCR |
| β-actin-R | CCTTGATGTCCCTGACGATT | RT-PCR |
| HMTLR4-sense | GGTCAGTGTTTCGATTGGCG | DsRNA synthesis |
| HMTLR4-anti | CCCAAGTTGTTGTTGCTGATGA | DsRNA synthesis |
| Time (h) | Concentrations (pg/mL) | ||||
|---|---|---|---|---|---|
| 24 °C + LPS | 24 °C + PBS | 32 °C + LPS | 32 °C + PBS | ||
| TNF-α | 6 | 720.17 ± 95.03 * | 368.70 ± 7.11 | 791.65 ± 71.13 † | 373.44 ± 30.98 |
| 48 | 746.63 ± 57.24 * | 382.12 ± 25.34 | 749.39 ± 58.73 † | 360.01 ± 30.10 | |
| LZM | 6 | 367.00 ± 54.31 * | 188.30 ± 24.78 | 424.84 ± 27.20 † | 240.54 ± 37.83 |
| 48 | 387.73 ± 38.14 * | 458.63 ± 36.42 | 458.63 ± 36.42 † | 214.01 ± 9.44 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, C.; Zhang, K.; Liu, Z.; Lü, W.; Guo, H.; Zhao, L.; Song, X.; Fang, J.K.-H. The Role of the TLR4-MyD88 Signaling Pathway in the Immune Response of the Selected Scallop Strain “Hongmo No. 1” to Heat Stress. Animals 2024, 14, 497. https://doi.org/10.3390/ani14030497
Yue C, Zhang K, Liu Z, Lü W, Guo H, Zhao L, Song X, Fang JK-H. The Role of the TLR4-MyD88 Signaling Pathway in the Immune Response of the Selected Scallop Strain “Hongmo No. 1” to Heat Stress. Animals. 2024; 14(3):497. https://doi.org/10.3390/ani14030497
Chicago/Turabian StyleYue, Chenyang, Kexin Zhang, Zhigang Liu, Wengang Lü, Hui Guo, Liqiang Zhao, Xinyu Song, and James Kar-Hei Fang. 2024. "The Role of the TLR4-MyD88 Signaling Pathway in the Immune Response of the Selected Scallop Strain “Hongmo No. 1” to Heat Stress" Animals 14, no. 3: 497. https://doi.org/10.3390/ani14030497





