Adaptive Evolution of the Greater Horseshoe Bat AANAT: Insights into the Link between AANAT and Hibernation Rhythms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Amplification, Cloning, and Sequencing
2.3. Recombinant Protein Preparation
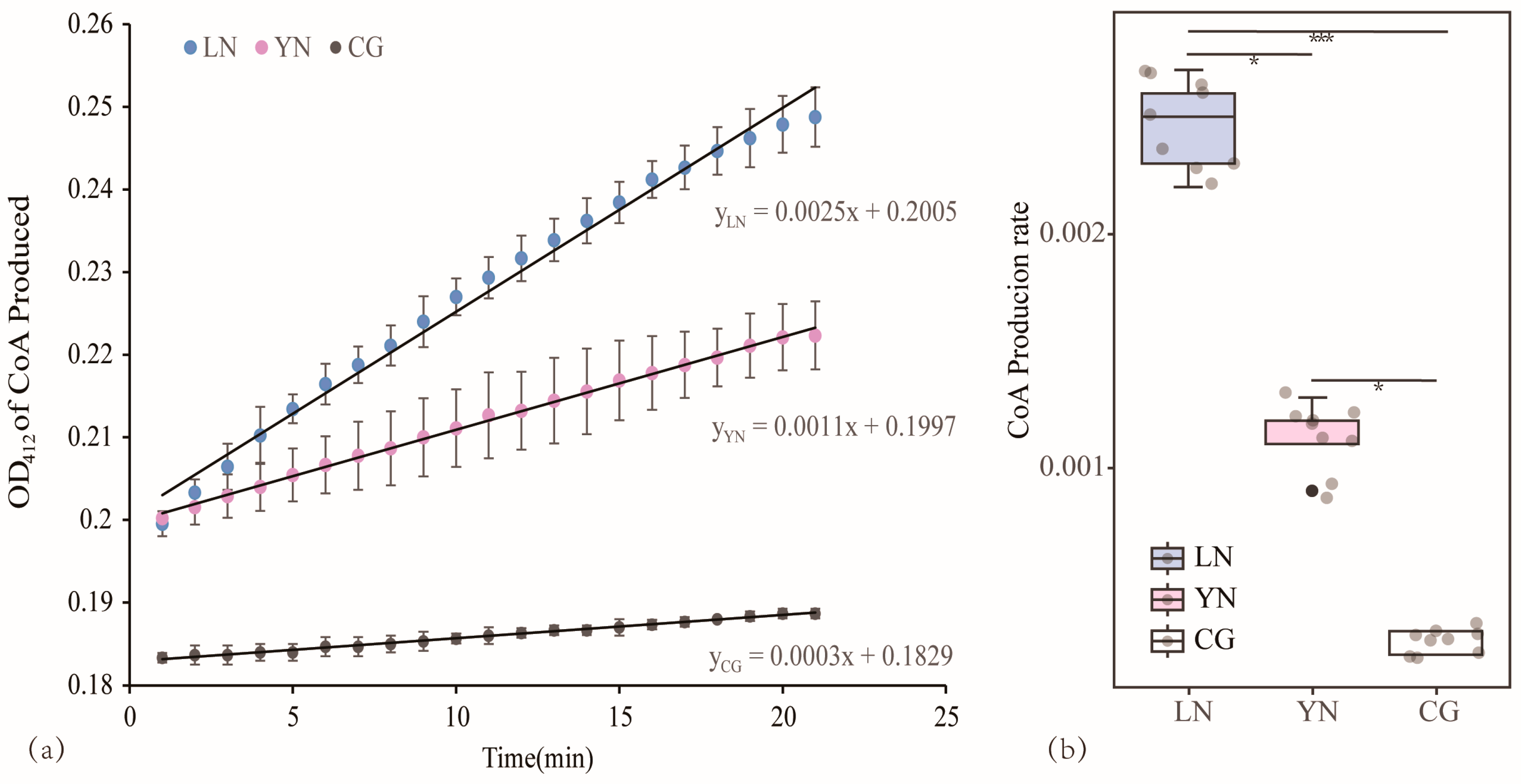
2.4. Enzymatic Activity Assays
2.5. Analysis of AANAT Gene Polymorphism
2.6. Analysis of Selective Pressure
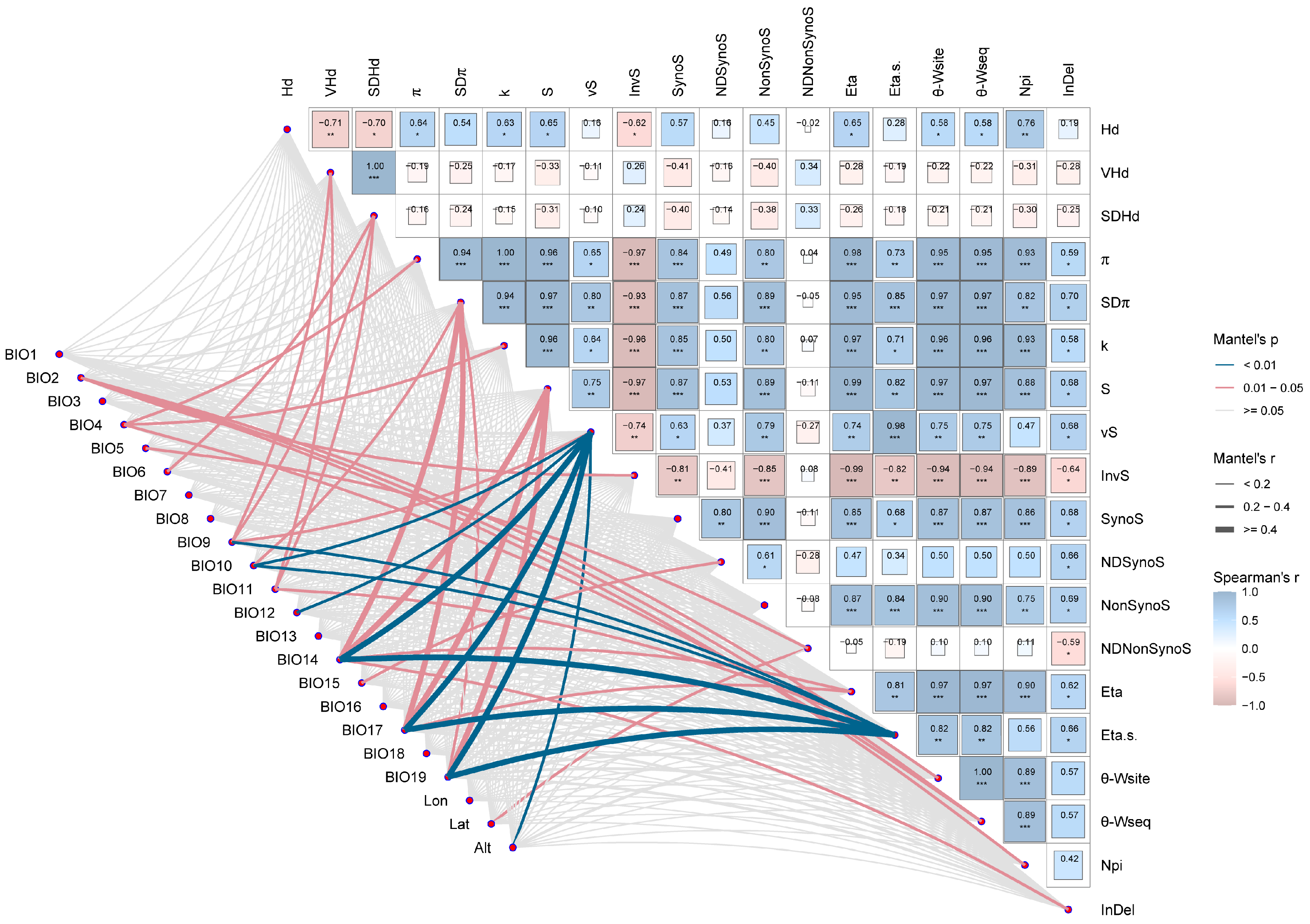
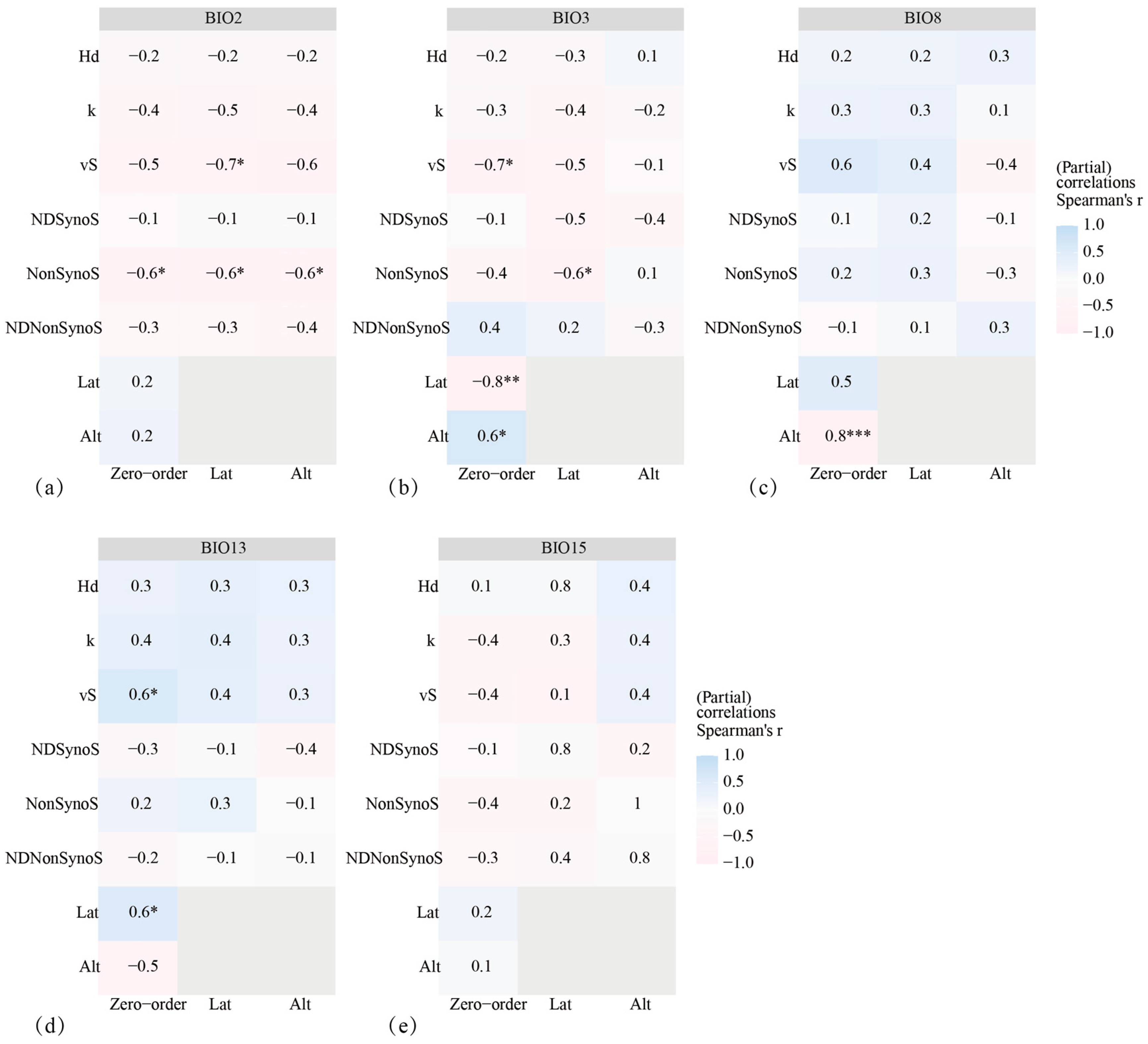
2.7. Effects of Environmental Factors on AANAT Polymorphism
3. Results
3.1. AANAT Gene Characteristics
3.2. AANAT Phylogeny
3.3. Selective Pressure on AANAT
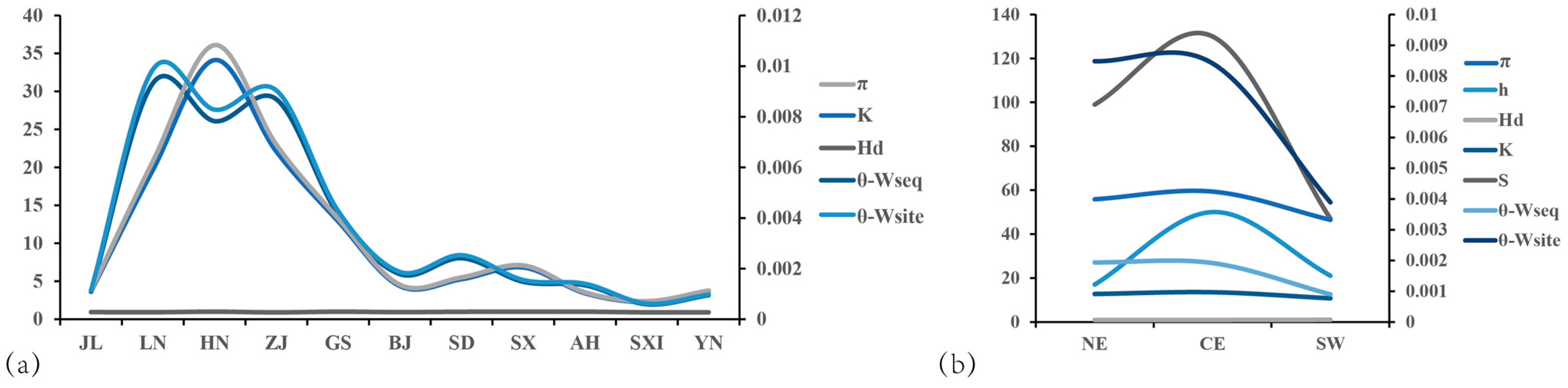
3.4. Comparison of the Catalytic Capacity of AANAT in Various Geographic Populations
3.5. Prediction of the AANAT Protein Structure
3.6. Drivers of Adaptive Evolution in AANAT
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hastings, M.H.; Reddy, A.B.; Maywood, E.S. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 2003, 4, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Wikelski, M.; Martin, L.B.; Scheuerlein, A.; Robinson, M.T.; Robinson, N.D.; Helm, B.; Hau, M.; Gwinner, E. Avian circannual clocks: Adaptive significance and possible involvement of energy turnover in their proximate control. Philos. Trans. R. Soc. B-Biol. Sci. 2007, 363, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J. The melatonin rhythm: Both a clock and a calendar. Experientia 1993, 49, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Voultsios, A.; Kennaway, D.J.; Dawson, D. Salivary melatonin as a circadian phase marker: Validation and comparison to plasma melatonin. J. Biol. Rhythms 1997, 12, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.W.; Dabrosin, C.; Yin, X.; Fuster, M.M.; Arreola, A.; Rathmell, W.K.; Generali, D.; Nagaraju, G.P.; El-Rayes, B.; Ribatti, D.; et al. Broad targeting of angiogenesis for cancer prevention and therapy. Semin. Cancer Biol. 2015, 35, S224–S243. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.C.; Weller, J.L. Indole metabolism in the pineal gland: A circadian rhythm in N-acetyltransferase. Science 1970, 169, 1093–1095. [Google Scholar] [CrossRef]
- Klein, D.C.; Moore, R.Y. Pineal N-acetyltransferase and hydroxyindole-O-methyl-transferase: Control by the retinohypothalamic tract and the suprachiasmatic nucleus. Brain Res. 1979, 174, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.; Mummaneni, P.; Steinbach, P.J.; Klein, D.C.; Coon, S.L. Characterization of the Saccharomyces cerevisiae homolog of the melatonin rhythm enzyme arylalkylamine N-acetyltransferase (EC 2.3. 1.87). J. Biol. Chem. 2001, 276, 47239–47247. [Google Scholar] [CrossRef]
- Falcón, J.; Coon, S.L.; Besseau, L.; Cazaméa-Catalan, D.; Fuentès, M.; Magnanou, E.; Paulin, C.H.; Boeuf, G.; Sauzet, S.; Jorgensen, E.H.; et al. Drastic neofunctionalization associated with evolution of the timezyme AANAT 500 Mya. Proc. Natl. Acad. Sci. USA 2014, 111, 314–319. [Google Scholar] [CrossRef]
- Pavlicek, J.; Sauzet, S.; Besseau, L.; Coon, S.L.; Weller, J.L.; Boeuf, G.; Gaildrat, P.; Omelchenko, M.V.; Koonin, E.V.; Falcón, J.; et al. Evolution of AANAT: Expansion of the gene family in the cephalochordate amphioxus. BMC Evol. Biol. 2010, 10, 154. [Google Scholar] [CrossRef]
- Cox, K.H.; Takahashi, J.S. Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 2019, 63, R93–R102. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, A.V.; Mosser, E.A.; Oikonomou, G.; Prober, D.A. Melatonin is required for the circadian regulation of sleep. Neuron 2015, 85, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, T.; Ma, X.; Wang, Y.; Liu, J.; Li, G.; Liu, Y.; Ji, P.; Zhang, Z.; Zhang, L.; et al. Melatonergic systems of AANAT, melatonin, and its receptor MT2 in the corpus luteum are essential for reproductive success in mammals. Biol. Reprod. 2021, 104, 430–444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Clough, S.J.; Adamah-Biassi, E.B.; Sveinsson, M.H.; Hutchinson, A.J.; Miura, I.; Furuse, T.; Wakana, S.; Matsumoto, Y.K.; Okanoya, K.; et al. Impact of endogenous melatonin on rhythmic behaviors, reproduction, and survival revealed in melatonin-proficient C57BL/6J congenic mice. J. Pineal Res. 2021, 71, e12748. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.Z.; Hallenbeck, J.M.; Cai, D.; McCarron, R.M. Elevated arylalkylamine-N-acetyltransferase (AA-NAT) gene expression in medial habenular and suprachiasmatic nuclei of hibernating ground squirrels. Mol. Brain Res. 2002, 102, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Stanton, T.L.; Daley, J.C.; Salzman, S.K. Prolongation of hibernation bout duration by continuous intracerebroventricular infusion of melatonin in hibernating ground squirrels. Brain Res. 1987, 413, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Mustonen, A.M.; Nieminen, P.; Puukka, M.; Asikainen, J.; Saarela, S.; Karonen, S.L.; Kukkonen, J.V.K.; Hyvarinen, H. Physiological adaptations of the raccoon dog (Nyctereutes procyonoides) to seasonal fasting-fat and nitrogen metabolism and influence of continuous melatonin treatment. J. Comp. Physiol. B 2004, 174, 1–12. [Google Scholar] [PubMed]
- Aboalroub, A.A.; Zhang, Z.; Keramisanou, D.; Gelis, I. Backbone resonance assignment of an insect arylalkylamine N-acetyltransferase from Bombyx mori reveals conformational heterogeneity. Biomol. NMR Assign. 2017, 11, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Zilberman-Peled, B.; Benhar, I.; Coon, S.L.; Ron, B.; Gothilf, Y. Duality of serotonin-N-acetyltransferase in the gilthead seabream (Sparus aurata): Molecular cloning and characterization of recombinant enzymes. Gen. Comp. Endocrinol. 2004, 138, 139–147. [Google Scholar] [CrossRef]
- Kim, W.; Park, H.; Seo, S. Global metabolic reconstruction and metabolic gene evolution in the cattle genome. PLoS ONE 2016, 11, e0150974. [Google Scholar] [CrossRef]
- Huelsmann, M.; Hecker, N.; Springer, M.S.; Gatesy, J.; Sharma, V.; Hiller, M. Genes lost during the transition from land to water in cetaceans highlight genomic changes associated with aquatic adaptations. Sci. Adv. 2019, 5, eaaw6671. [Google Scholar] [CrossRef]
- Valente, R.; Alves, F.; Sousa-Pinto, I.; Ruivo, R.; Castro, L.F.C. Functional or vestigial? The genomics of the pineal gland in Xenarthra. J. Mol. Evol. 2021, 89, 565–575. [Google Scholar] [CrossRef]
- Emerling, C.A.; Springer, M.S.; Gatesy, J.; Jones, Z.; Hamilton, D.; Xia-Zhu, D.; Collin, M.; Delsuc, F. Genomic evidence for the parallel regression of melatonin synthesis and signaling pathways in placental mammals [version 2; peer review: 2 approved]. Open Res. Eur. 2021, 1, 75. [Google Scholar] [CrossRef]
- Yin, D.Q.; Zhou, R.R.; Yin, M.X.; Chen, Y.; Xu, S.X.; Yang, G. Gene duplication and loss of AANAT in mammals driven by rhythmic adaptations. Mol. Biol. Evol. 2021, 38, 3925–3937. [Google Scholar] [CrossRef] [PubMed]
- Teeling, E.C.; Vernes, S.C.; Dávalos, L.M.; Ray, D.A.; Gilbert, M.T.P.; Myers, E. Bat biology, genomes, and the Bat1K project: To generate chromosome-level genomes for all living bat species. Annu. Rev. Anim. Biosci. 2018, 6, 23–46. [Google Scholar] [CrossRef]
- Sadier, A.; Urban, D.J.; Anthwal, N.; Howenstine, A.O.; Sinha, I.; Sears, K.E. Making a bat: The developmental basis of bat evolution. Genet. Mol. Biol. 2020, 43, e20190146. [Google Scholar] [CrossRef]
- Iuvone, P.M.; Tosini, G.; Pozdeyev, N.; Haque, R.; Klein, D.C.; Chaurasia, S.S. Circadian clocks, clock networks, arylalkylamine N-acetyltransferase, and melatonin in the retina. Prog. Retin. Eye Res. 2005, 24, 433–456. [Google Scholar] [CrossRef]
- Reiter, R.J. Pineal melatonin: Cell biology of its synthesis and of its physiological interactions. Endocr. Rev. 1991, 12, 151–180. [Google Scholar] [CrossRef] [PubMed]
- Haldar, C.; Yadav, R.; Alipreeta. Annual reproductive synchronization in ovary and pineal gland function of female short-nosed fruit bat, Cynopterus sphinx. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2006, 144, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.K.; Krishna, A. Melatonin modulates glucose homeostasis during winter dormancy in a vespertilionid bat, Scotophilus heathi. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2010, 155, 392–400. [Google Scholar] [CrossRef]
- Banerjee, A.; Udin, S.; Krishna, A. Regulation of leptin synthesis in white adipose tissue of the female fruit bat, Cynopterus sphinx role of melatonin with or without insulin. Exp. Physiol. 2011, 96, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.P.; Luo, L.; Kimball, R.T.; Wei, X.W.; Jin, L.R.; Jiang, T.L.; Li, G.H.; Feng, J. Geographic variation in the acoustic traits of greater horseshoe bats: Testing the importance of drift and ecological selection in evolutionary processes. PLoS ONE 2013, 8, e70368. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.H.; Wu, Y.H.; Sun, K.P.; Wang, H.; Jiang, T.L.; Lin, A.Q.; Huang, X.B.; Yue, X.K.; Shi, L.M.; Feng, J. Gene expression and adaptive evolution of ZBED1 in the hibernating greater horseshoe bat (Rhinolophus ferrumequinum). J. Exp. Biol. 2016, 219, 834–843. [Google Scholar] [PubMed]
- Pavlicek, J.; Coon, S.L.; Ganguly, S.; Weller, J.L.; Hassan, S.A.; Sackett, D.L.; Klein, D.C. Evidence that proline focuses movement of the floppy loop of arylalkylamine N-acetyltransferase (EC 2.3.1.87). J. Biol. Chem. 2008, 283, 14552–14558. [Google Scholar] [CrossRef]
- Leng, P.Q.; Zhao, F.L.; Yin, B.C.; Ye, B.C. A novel, colorimetric method for biogenic amine detection based on arylalkylamine N-acetyltransferase. Chem. Commun. 2015, 51, 8712–8714. [Google Scholar] [CrossRef] [PubMed]
- Bolker, B.M.; Brooks, M.E.; Clark, C.J.; Geange, S.W.; Poulsen, J.R.; Stevens, M.H.H.; White, J.S.S. Generalized linear mixed models: A practical guide for ecology and evolution. Trends Ecol. Evol. 2009, 24, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Zhao, H.B. Molecular Mechanism of Geographic Variation Inecholocation Calls of the Greater Horseshoe Bat. Ph.D. Thesis, Northeast Normal University, Changchun, China, 2020. [Google Scholar]
- Yang, Z.H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Goodchild, M.F. Geographical data modeling. Comput. Geosci. 1992, 18, 401–408. [Google Scholar] [CrossRef]
- Francioli, L.C.; Menelaou, A.; Pulit, S.L.; Van Dijk, F.; Palamara, P.F.; Elbers, C.C.; Neerincx, P.B.T.; Ye, K.; Guryev, V.; Kloosterman, W.P.; et al. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014, 46, 818–825. [Google Scholar]
- Luo, L. Genetic Diversity and Population Structure of Rhinolophus ferrumequinum in China Based on Microsatellite. Master’s Thesis, Northeast Normal University, Changchun, China, 2011. [Google Scholar]
- Zhang, L.; Tang, Y.; Merkler, D.J.; Han, Q. Function, structure, evolution, regulation of a potent drug target, arylalkylamine N-acetyltransferase. Adv. Protein Chem. Struct. Biol. 2023, 134, 211–223. [Google Scholar] [PubMed]
- Hu, Y.B.; Fan, H.Z.; Chen, Y.H.; Chang, J.; Zhan, X.J.; Wu, H.; Zhang, B.W.; Wang, M.; Zhang, W.Y.; Yang, L.; et al. Spatial patterns and conservation of genetic and phylogenetic diversity of wildlife in China. Sci. Adv. 2021, 7, eabd5725. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Chen, H.Q.; Lin, Z.N.; Zhang, L.; Upadhyay, A.; Liao, C.H.; Merkler, D.J.; Han, Q. Evolutionary genomics analysis reveals gene expansion and functional diversity of arylalkylamine N-acetyltransferases in the Culicinae subfamily of mosquitoes. Insect Sci. 2023, 30, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Falcón, J.; Besseau, L.; Fuentès, M.; Sauzet, S.; Magnanou, E.; Boeuf, G. Structural and functional evolution of the pineal melatonin system in vertebrates. Ann. N. Y. Acad. Sci. 2009, 1163, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Hohjoh, H.; Takasu, M.; Shishikura, K.; Takahashi, Y.; Honda, Y.; Tokunaga, K. Significant association of the arylalkylamine N-acetyltransferase (AA-NAT) gene with delayed sleep phase syndrome. Neurogenetics 2003, 4, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Back, K. Low melatonin production by suppression of either serotonin N-acetyltransferase or N-acetylserotonin methyltransferase in rice causes seedling growth retardation with yield penalty, abiotic stress susceptibility, and enhanced coleoptile growth under anoxic conditions. J. Pineal Res. 2016, 60, 348–359. [Google Scholar] [PubMed]
- Mustonen, A.-M.; Nieminen, P. A review of the physiology of a survival expert of big freeze, deep snow, and an empty stomach: The boreal raccoon dog (Nyctereutes procyonoides). J. Comp. Physiol., B 2018, 188, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Nieminen, P.; Mustonen, A.-M.; Asikainen, J.; Hyvärinen, H. Seasonal weight regulation of the raccoon dog (Nyctereutes procyonoides): Interactions between melatonin, leptin, ghrelin, and growth hormone. J. Biol. Rhythms 2002, 17, 155–163. [Google Scholar] [CrossRef]
- Schwartz, C.; Ballinger, M.A.; Andrews, M.T. Melatonin receptor signaling contributes to neuroprotection upon arousal from torpor in thirteen-lined ground squirrels. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2015, 309, R1292–R1300. [Google Scholar] [CrossRef]
- Reiter, R.J.; Rosales-Corral, S.; Tan, D.X.; Jou, M.J.; Galano, A.; Xu, B. Melatonin as a mitochondria-targeted antioxidant: One of evolution’s best ideas. Cell. Mol. Life Sci. 2017, 74, 3863–3881. [Google Scholar] [CrossRef] [PubMed]
- Willis, C.K.R.; Wilcox, A. Hormones and hibernation: Possible links between hormone systems, winter energy balance and white-nose syndrome in bats. Horm. Behav. 2014, 66, 66–73. [Google Scholar] [CrossRef]
- McStay, E.; Migaud, H.; Vera, L.M.; Sánchez-Vázquez, F.J.; Davie, A. Comparative study of pineal clock gene and AANAT2 expression in relation to melatonin synthesis in Atlantic salmon (Salmo salar) and European seabass (Dicentrarchus labrax). Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2014, 169, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Ringo, J.L.; Doty, R.W.; Demeter, S.; Simard, P.Y. Time is of the essence a conjecture that hemispheric specialization arises from interhemispheric conduction delay. Cereb. Cortex 1994, 4, 331–343. [Google Scholar] [CrossRef] [PubMed]
- van Oort, B.E.H.; Tyler, N.J.C.; Gerkema, M.P.; Folkow, L.; Stokkan, K.-A. Where clocks are redundant: Weak circadian mechanisms in reindeer living under polar photic conditions. Naturwissenschaften 2007, 94, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Arnold, W.; Ruf, T.; Loe, L.E.; Irvine, R.J.; Ropstad, E.; Veiberg, V.; Albon, S.D. Circadian rhythmicity persists through the Polar night and midnight sun in Svalbard reindeer. Sci. Rep. 2018, 8, 14466. [Google Scholar] [CrossRef]
- Kerkhoff, A.J.; Moriarty, P.E.; Weiser, M.D. The latitudinal species richness gradient in New World woody angiosperms is consistent with the tropical conservatism hypothesis. Proc. Natl. Acad. Sci. USA 2014, 111, 8125–8130. [Google Scholar] [CrossRef]
- Sharma, V.; Hecker, N.; Roscito, J.G.; Foerster, L.; Langer, B.E.; Hiller, M. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 2018, 9, 1215. [Google Scholar] [CrossRef] [PubMed]
- Coon, S.L.; Klein, D.C. Evolution of arylalkylamine N-acetyltransferase: Emergence and divergence. Mol. Cell. Endocrinol. 2006, 252, 2–10. [Google Scholar] [CrossRef]
- Isorna, E.; Aliaga-Guerrero, M.; M’Rabet, A.E.; Servili, A.; Falcón, J.; Muñoz-Cueto, J.A. Identification of two arylalkylamine N-acetyltranferase1 genes with different developmental expression profiles in the flatfish Solea senegalensis. J. Pineal Res. 2011, 51, 434–444. [Google Scholar] [CrossRef]
- Paulin, C.H.; Cazaméa-Catalan, D.; Zilberman-Peled, B.; Herrera-Perez, P.; Sauzet, S.; Magnanou, E.; Fuentès, M.; Gothilf, Y.; Muñoz-Cueto, J.A.; Falcón, J.; et al. Subfunctionalization of arylalkylamine N-acetyltransferases in the sea bass Dicentrarchus labrax two-ones for one two. J. Pineal Res. 2015, 59, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Lyamin, O.I.; Manger, P.R.; Ridgway, S.H.; Mukhametov, L.M.; Siegel, J.M. Cetacean sleep: An unusual form of mammalian sleep. Neurosci. Biobehav. Rev. 2008, 32, 1451–1484. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Aravind, L.; Coon, S.L.; Klein, D.C.; Koonin, E.V. Evolution of cell-cell signaling in animals: Did late horizontal gene transfer from bacteria have a role? Trends Genet. 2004, 20, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenberger, A.; Handke, N.H.; Romer, T.; Wacker, A. Geographic clines in Daphnia magna’s circadian clock gene expression: Local adaptation to photoperiod. Zoology 2021, 144, 12856. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.M.; Jesus, T.F.; Coelho, M.M.; Sousa, V.C. Adaptation and convergence in circadian-related genes in Iberian freshwater fish. BMC Ecol. Evol. 2021, 21, 38. [Google Scholar] [CrossRef] [PubMed]
- McCain, C.M.; King, S.R.B. Body size and activity times mediate mammalian responses to climate change. Glob. Chang. Biol. 2014, 20, 1760–1769. [Google Scholar] [CrossRef]
- Li, X.L.; Liu, T.; Li, A.Q.; Xiao, Y.H.; Sun, K.P.; Feng, J. Diversifying selection and climatic effects on major histocompatibility complex class II gene diversity in the greater horseshoe bat. Evol. Appl. 2023, 16, 688–704. [Google Scholar] [CrossRef]



| Name | Base Sequence (5′-3′) | Annealing | Extending | Products |
|---|---|---|---|---|
| 1.1F | TCAACACCTAGCAAGAGC | 55 °C | 2 min | 1981 bp |
| 1.1R | GTCCCAAAGTGAACCGAT | |||
| 1.2F | CTGTTACCTGCGGCTCAAC | 57 °C | 2.5 min | 2394 bp |
| 1.2R | TGTCACCTCTGCGGATACCT | |||
| 2F | TGCCGCAGCCTTCATCTCTGTCTCG | 57 °C | 1.8 min | 1712 bp |
| 2R | CCTCAGGAAGAATGAAAGCTGGAACCTT |
| Abbreviation | Bioclimatic Variable |
|---|---|
| BIO1 | Annual Mean Temperature |
| BIO2 | Monthly Mean Temperature |
| BIO3 | Isothermality (Bio2/Bio7) (×100) |
| BIO4 | Temperature Seasonality (standard deviation) (×100) |
| BIO5 | Max Temperature of Warmest Month |
| BIO6 | Min Temperature of Coldest Month |
| BIO7 | Temperature Annual Range (Bio5-Bio6) |
| BIO8 | Mean Temperature of Wettest Quarter |
| BIO9 | Mean Temperature of Driest Quarter |
| BIO10 | Mean Temperature of Warmest Quarter |
| BIO11 | Mean Temperature of Coldest Quarter |
| BIO12 | Annual Precipitation |
| BIO13 | Precipitation of Wettest Month |
| BIO14 | Precipitation of Driest Month |
| BIO15 | Precipitation Seasonality (Coefficient of Variation) |
| BIO16 | Precipitation of Wettest Quarter |
| BIO17 | Precipitation of Driest Quarter |
| BIO18 | Precipitation of Warmest Quarter |
| BIO19 | Precipitation of Coldest Quarter |
| Model | Parameters Estimate | Ln L | df | 2ΔLn L | p-Value | Positively Selected Sites | |
|---|---|---|---|---|---|---|---|
| M0 (one-ratio) | ω = 0.05305 | −1010.615442 | 29 | None | |||
| M1a (neutral) | p0 = 0.95691, p1 = 0.04309 ω0 = 0.00000, ω1 = 1.00000 | −1002.789947 | 30 | None | |||
| M2a (selection) | p0 = 0.97461, p1 = 0.00000, p2 = 0.02539 ω0 = 0.00490, ω1 = 1.00000, ω2 = 1.93757 | −1002.481582 | 32 | 70I (0.573), 187A (0.800) | |||
| M3 (discrete) | p0 = 0.29208, p1 = 0.68253, p2 = 0.02539 ω0 = 0.00490, ω1 = 0.00490, ω2 = 1.93760 | −1002.481582 | 33 | None | |||
| M7 (beta) | p = 0.01000, q = 0.17324 | −1004.096785 | 30 | None | |||
| M8 (beta and ω > 1) | p0 = 0.97460, p = 0.51911, q = 99.00000 (p1 = 0.02540), ω = 1.93628 | −1002.482059 | 32 | 70I (0.693), 187A (0.906) | |||
| M8a (beta and ω = 1) | p0 = 0.95691, p = 0.00500, q = 2.54319 (p1 = 0.04309), ω = 1.00000 | −1002.789924 | 31 | None | |||
| LRT of variable ω values among branches | |||||||
| M0 vs. M3 | 4 | 16.26772 | 0.05 < p < 0.1 | ||||
| M1a vs. M2a | 2 | 0.61673 | 0.5 < p < 0.9 | ||||
| M7 vs. M8 | 2 | 3.229452 | 0.1 < p < 0.5 | ||||
| M8 vs. M8a | 1 | 0.61573 | 0.1 < p < 0.5 | ||||
| Model | Foreground Branch | Parameter Estimates | Ln L | df | 2ΔLn L | p-Value | |
|---|---|---|---|---|---|---|---|
| One-ratio | ω = 0.05304 | −1010.615442 | 30 | ||||
| Free-ratio | −1009.141884 | 57 | |||||
| Two-ratio | NE | ω0 = 0.05688, ω1 = 0.04539 | −1010.584015 | 31 | |||
| CE | ω0 = 0.04230, ω1 = 0.05905 | −1010.545700 | 31 | ||||
| SW | ω0 = 0.05438, ω1 = 0.00010 | −1010.459894 | 31 | ||||
| NE + CE | ω0 = 0.00010, ω1 = 0.05438 | −1010.459894 | 31 | ||||
| Two-ratio, ω = 1 | NE | ω0 = 0.05686, ω1 = 1.00000 | −1024.101938 | 30 | |||
| CE | ω0 = 0.04229, ω1 = 1.00000 | −1034.724656 | 30 | ||||
| SW | ω0 = 0.05305, ω1 = 1.00000 | −1010.615980 | 30 | ||||
| NE + CE | ω0 = 0.00010, ω1 = 1.00000 | −1048.017833 | 30 | ||||
| LRT of variable ω values among branches | |||||||
| Free-ratio vs. one-ratio | 27 | 2.947116 | 1 | ||||
| LRT of ω at specific lineages | |||||||
| Two-ratio vs. one-ratio | NE | 1 | 0.062854 | 0.5 < p < 0.9 | |||
| CE | 1 | 0.139484 | 0.5 < p < 0.9 | ||||
| SW | 1 | 0.311096 | 0.5 < p < 0.9 | ||||
| NE + CE | 1 | 0.311096 | 0.5 < p < 0.9 | ||||
| Two-ratio vs. two-ratio (ω1 = 1) | NE | 1 | 27.035846 | p < 0.001 | |||
| CE | 1 | 48.357912 | p < 0.001 | ||||
| SW | 1 | 0.312172 | 0.5 < p < 0.9 | ||||
| NE + CE | 1 | 75.115878 | p < 0.001 | ||||
| Foreground Branch | Parameter Estimates | Ln L | df | 2ΔLn L | np | p-Value | Selected Sites |
|---|---|---|---|---|---|---|---|
| NE | Null | −1001.926974 | 1 | 0.134988 | 32 | 0.9 < p < 0.97 | None |
| p0 = 0.96733, p1 = 0.03267, p2a = 0.00000, p2b = 0.00000 | |||||||
| Background: ω0 = 0.00000, ω1 = 1.00000, ω2a = 0.00000, ω2b = 1.00000 | |||||||
| Foreground: ω0 = 0.00000, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 | |||||||
| Aternative | −1001.859480 | 33 | |||||
| p0 = 0.96160, p1 = 0.02817, p2a = 0.00994, p2b = 0.00029 | |||||||
| Background: ω0 = 0.00000, ω1 = 1.00000, ω2a = 0.00000, ω2b = 1.00000 | |||||||
| Foreground: ω0 = 0.00000, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 | |||||||
| CE | Null | −1001.926974 | 1 | 0 | 32 | 1 | None |
| p0 = 0.96733, p1 = 0.03267, p2a = 0.00000, p2b = 0.00000 | |||||||
| Background: ω0 = 0.00000, ω1 = 1.00000, ω2a = 0.00000, ω2b = 1.00000 | |||||||
| Foreground: ω0 = 0.00000, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 | |||||||
| Aternative | −1001.926974 | 33 | |||||
| p0 = 0.96733, p1 = 0.03267, p2a = 0.00000, p2b = 0.00000 | |||||||
| Background: ω0 = 0.00000, ω1 = 1.00000, ω2a = 0.00000, ω2b = 1.00000 | |||||||
| Foreground: ω0 = 0.00000, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 | |||||||
| NE + CE | Null | −1002.650389 | 1 | 0.666436 | 32 | 0.1 < p < 0.5 | 70I (0.587) 187A (0.811) |
| p0 = 0.95622, p1 = 0.00000, p2a = 0.04378, p2b = 0.00000 | |||||||
| Background: ω0 = 0.00000, ω1 = 1.00000, ω2a = 0.00000, ω2b = 1.00000 | |||||||
| Foreground: ω0 = 0.00000, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 | |||||||
| Aternative | −1002.317171 | 33 | |||||
| p0 = 0.97334, p1 = 0.00000, p2a = 0.02666, p2b = 0.00000 | |||||||
| Background: ω0 = 0.00396, ω1 = 1.00000, ω2a = 0.00396, ω2b = 1.00000 | |||||||
| Foreground: ω0 = 0.00396, ω1 = 1.00000, ω2a = 1.93612, ω2b = 1.93612 | |||||||
| SW | Null | −1002.789947 | 1 | 0 | 32 | 1 | None |
| p0 = 0.95691, p1 = 0.04309, p2a = 0.00000, p2b = 0.00000 | |||||||
| Background: ω0 = 0.00000, ω1 = 1.00000, ω2a = 0.00000, ω2b = 1.00000 | |||||||
| Foreground: ω0 = 0.00000, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 | |||||||
| Aternative | −1002.789947 | 33 | |||||
| p0 = 0.95691, p1 = 0.04309, p2a = 0.00000, p2b = 0.00000 | |||||||
| Background: ω0 = 0.00000, ω1 = 1.00000, ω2a = 0.00000, ω2b = 1.00000 | |||||||
| Foreground: ω0 = 0.00000, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 |
| Fixed Effects | Estimate | Std. Error | t-Value | p-Value |
|---|---|---|---|---|
| Intercept 1 | −8.23400 | 0.05266 | −156.35 | <0.001 *** |
| LN vs. CG | 2.23374 | 0.06129 | 36.45 | <0.001 *** |
| YN vs. CG | 1.44237 | 0.06141 | 23.49 | <0.001 *** |
| Intercept 2 | −6.00039 | 0.04563 | −131.51 | <0.001 *** |
| YN vs. LN | −0.79205 | 0.06452 | −12.28 | <0.001 *** |
| CG vs. LN | −2.22912 | 0.06452 | −34.55 | <0.001 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Wang, L.; Liu, S.; Pu, Y.; Sun, K.; Xiao, Y.; Feng, J. Adaptive Evolution of the Greater Horseshoe Bat AANAT: Insights into the Link between AANAT and Hibernation Rhythms. Animals 2024, 14, 1426. https://doi.org/10.3390/ani14101426
Zhao Y, Wang L, Liu S, Pu Y, Sun K, Xiao Y, Feng J. Adaptive Evolution of the Greater Horseshoe Bat AANAT: Insights into the Link between AANAT and Hibernation Rhythms. Animals. 2024; 14(10):1426. https://doi.org/10.3390/ani14101426
Chicago/Turabian StyleZhao, Yanhui, Lei Wang, Sen Liu, Yingting Pu, Keping Sun, Yanhong Xiao, and Jiang Feng. 2024. "Adaptive Evolution of the Greater Horseshoe Bat AANAT: Insights into the Link between AANAT and Hibernation Rhythms" Animals 14, no. 10: 1426. https://doi.org/10.3390/ani14101426





