Chromosome Diversity and Evolution of the Endemic Malagasy Velvet Geckos of the Genus Blaesodactylus (Reptilia, Gekkonidae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling
2.2. Molecular and Phylogenetic Analysis
2.3. Cytogenetic Analysis
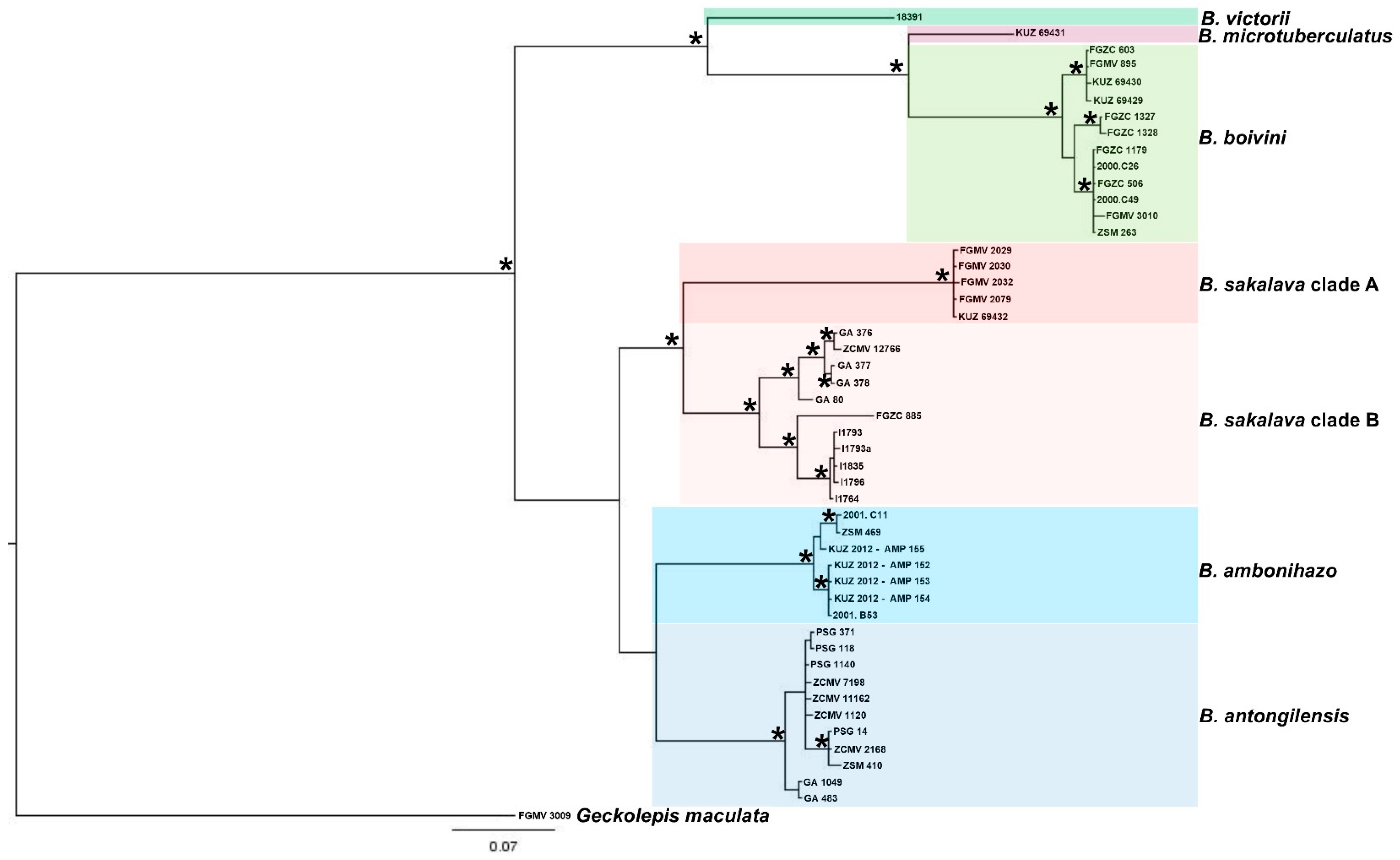
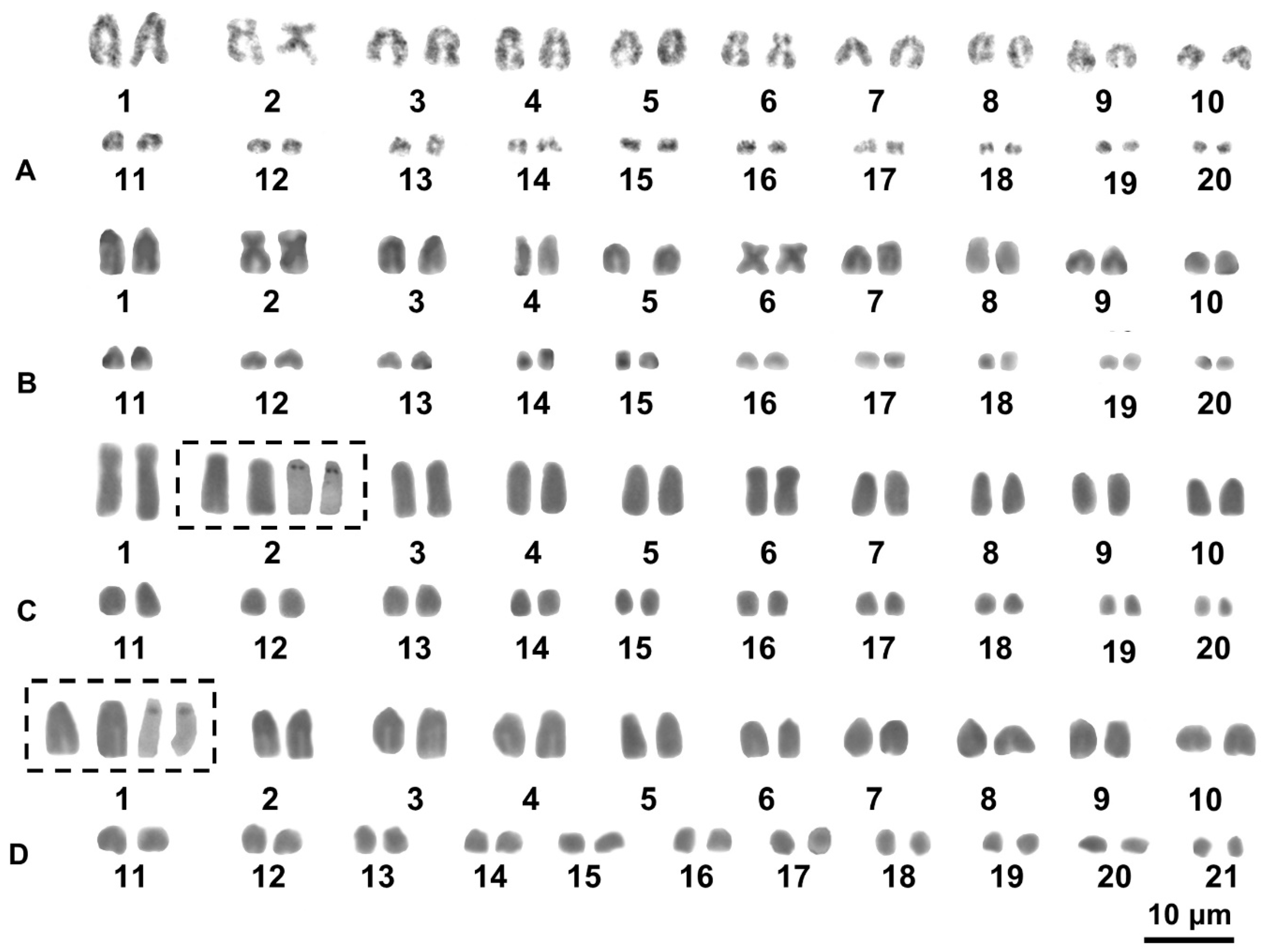
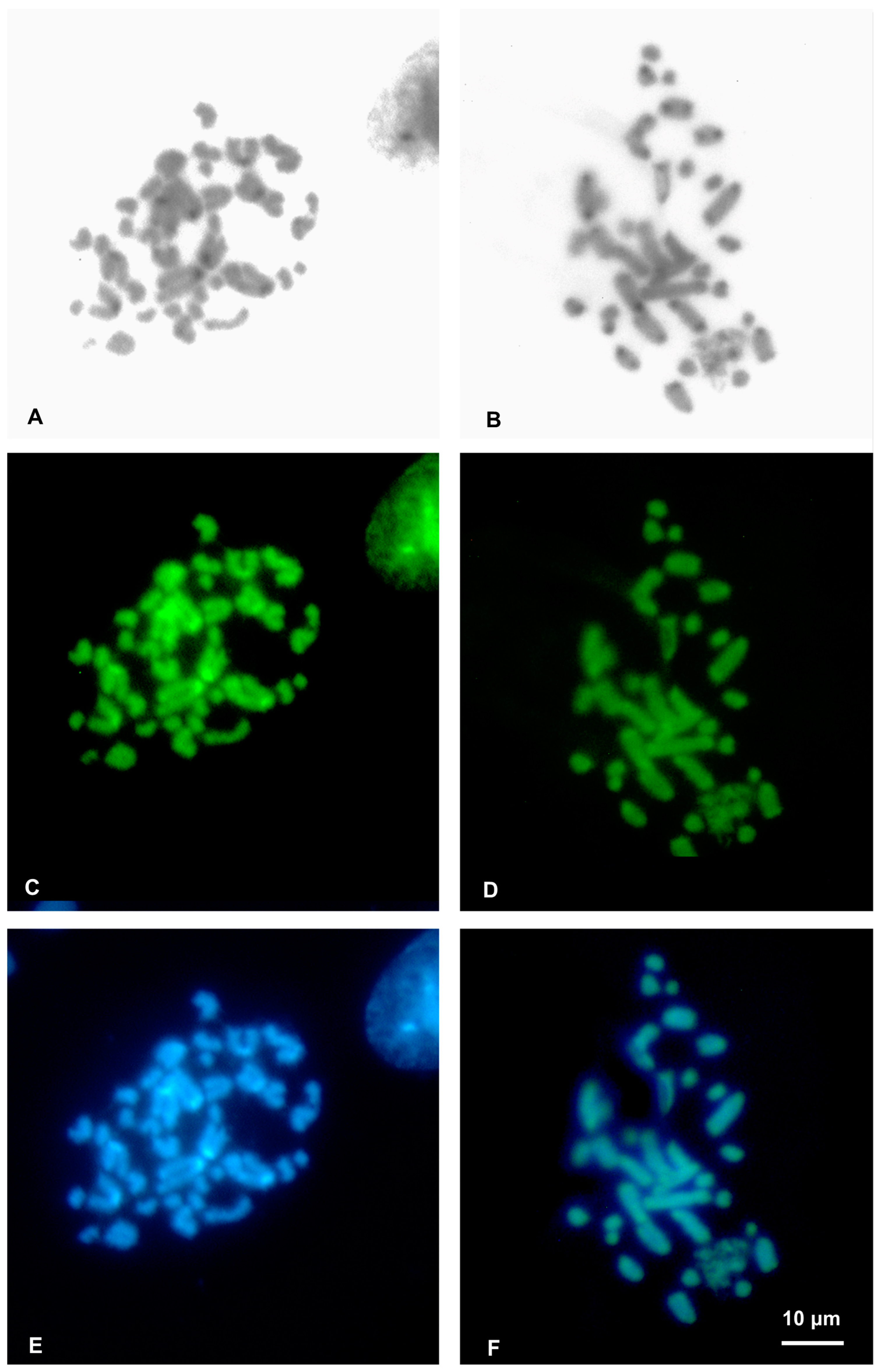
3. Results
3.1. Molecular and Phylogenetic Analysis
3.2. Cytogenetic Analysis
4. Discussion
4.1. Molecular Analysis
4.2. Cytogenetic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hot-spots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Ganzhorn, J.U.; Lowry II, P.P.; Schatz, G.E.; Somme, S. The biodiversity of Madagascar: One of the world’s hottest hotspots on its way out. Oryx 2001, 35, 346–348. [Google Scholar] [CrossRef]
- Vences, M.; Wollenberg, K.C.; Vieites, D.R.; Lees, D.C. Madagascar as a model region of species diversification. Trends Ecol. Evol. 2009, 24, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Glaw, F.; Vences, M. A Field Guide to the Amphibians and Reptiles of Madagascar, 3rd ed.; Verlags GbR: Köln, Germany, 2007; p. 496. [Google Scholar]
- Uetz, P.; Freed, P.; Hošek, J. The Reptile Database. 2022. Available online: http://www.reptile-database.org (accessed on 22 March 2023).
- Koubová, M.; Pokorná, M.J.; Rovatsos, M.; Farkacová, K.; Altmanová, M.; Kratochvíl, L. Sex determination in Madagascar geckosof the genus Paroedura (Squamata: Gekkonidae): Are differentiated sex chromosomes indeed so evolutionary stable? Chromosom. Res. 2014, 22, 441–452. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Glaw, F.; Odierna, G.; Petraccioli, A.; Guarino, F.M. Chromosome aneupolyploidy in an endemic Malagasy gecko (Gekkonidae: Geckolepis). Salamandra 2018, 54, 56–62. [Google Scholar]
- Mezzasalma, M.; Andreone, F.; Glaw, F.; Guarino, F.M.; Odierna, G.; Petraccioli, A.; Picariello, O. Changes in heterochromatin content and ancient chromosome fusion in the endemic Malagasy boid snakes Sanzinia and Acrantophis (Squamata: Serpentes). Salamandra 2019, 55, 140–144. [Google Scholar]
- Mezzasalma, M.; Brunelli, E.; Odierna, G.; Guarino, F.M. First Insights on the Karyotype Diversification of the Endemic Malagasy Leaf-Toed Geckos (Squamata: Gekkonidae: Uroplatus). Animals 2022, 12, 2054. [Google Scholar] [CrossRef]
- Ayala, F.J.; Coluzzi, M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Natl. Acad. Sci. USA 2005, 102, 6535–6542. [Google Scholar] [CrossRef] [Green Version]
- Faria, R.; Navarro, A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol. Evol. 2010, 25, 660–669. [Google Scholar] [CrossRef]
- Olmo, E. Trends in the evolution of reptilian chromosomes. Integr. Comp. Biol. 2008, 48, 486–493. [Google Scholar] [CrossRef]
- Petraccioli, A.; Guarino, F.M.; Kupriyanova, L.; Mezzasalma, M.; Odierna, G.; Picariello, O.; Capriglione, T. Isolation and characterization of interspersed repeated sequences in the common lizard, Zootoca vivipara, and their conservation in squamata. Cytogenet. Genome Res. 2019, 157, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Sidhom, M.; Said, K.; Chatti, N.; Guarino, F.M.; Odierna, G.; Petraccioli, A.; Picariello, O.; Mezzasalma, M. Karyological characterization of the common chameleon (Chamaeleo chamaeleon) provides insights on the evolution and diversification of sex chromosomes in Chamaeleonidae. Zoology 2020, 141, 125738. [Google Scholar] [CrossRef] [PubMed]
- Mezzasalma, M.; Guarino, F.M.; Odierna, G. Lizards as Model Organisms of Sex Chromosome Evolution: What We Really Know from a Systematic Distribution of Available Data? Genes 2021, 12, 1341. [Google Scholar] [CrossRef]
- Greembaum, E.; Bauer, A.M.; Jackmann, T.R. Homopholis and Blaesodactylus (Squamata: Gekkonidae) revisited new insights from a molecular phylogeny. Afr. J. Herpetol. 2007, 56, 101–114. [Google Scholar] [CrossRef]
- Bauer, A.M.; Glaw, F.; Gehring, P.-S.; Vences, M. New species of Blaesodactylus (Squamata: Gekkonidae) from Ankarafantsika National Park in north-western Madagascar. Zootaxa 2011, 2942, 57–68. [Google Scholar] [CrossRef]
- Jono, T.; Bauer, A.M.; Brennan, I.; Mori, A. New species of Blaesodactylus (Squamata: Gekkonidae) from Tsingy karstic outcrops in Ankarana National Park, northern Madagascar. Zootaxa 2015, 3980, 406–416. [Google Scholar] [CrossRef] [Green Version]
- Ineich, I.; Glaw, F.; Vences, M. A new species of Blaesodactylus (Squamata: Gekkonidae) from Tsingy limestone outcrops in Namoroka National Park, north-western Madagascar. Zootaxa 2016, 4109, 523–541. [Google Scholar] [CrossRef]
- Chrostek, G.; Domaradzka, A.; Yurchenko, A.; Kratochvíl, L.; Mazzoleni, S.; Rovatsos, M. Cytogenetic Analysis of Seven Species of Gekkonid and Phyllodactylid Geckos. Genes 2023, 14, 178. [Google Scholar] [CrossRef]
- Jacobsen, N.H.; Kuhn, A.L.; Kuhn, A.L.; Jackman, T.R.; Bauer, A.M. A phylogenetic analysis of the southern African gecko genus Afroedura Loveridge (Squamata: Gekkonidae), with the description of nine new species from Limpopo and Mpumalanga provinces of South Africa. Zootaxa 2014, 3846, 451–501. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Lab Press: New York, NY, USA, 1989. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Mezzasalma, M.; Brunelli, E.; Odierna, G.; Guarino, F.M. Comparative cytogenetics of Hemorrhois hippocrepis and Malpolon monspessulanus highlights divergent karyotypes in Colubridae and Psammophiidae (Squamata: Serpentes). Eur. Zool. J. 2023, 90, 201–210. [Google Scholar] [CrossRef]
- Howell, W.M.; Black, D.A. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: A1-step method. Experientia 1980, 36, 1014–1015. [Google Scholar] [CrossRef] [PubMed]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- De Smet, W.H.O. Description of the orcien stained karyotypes of 27 lizard species (Lacertilia: Reptilia) belonging to the families Iguanidae, Agamidae Chameleontidae and Gekkonidae (Ascalabota). Acta Zool. Pathol. Antverpiensia 1981, 76, 35–72. [Google Scholar]
- Rovatsos, M.; Kratochvíl, L.; Altmanová, M.; Johnson Pokorná, M. Interstitial Telomeric Motifs in Squamate Reptiles: When the Exceptions Outnumber the Rule. PLoS ONE 2015, 10, e0134985. [Google Scholar] [CrossRef] [Green Version]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 93. [Google Scholar] [CrossRef] [Green Version]
- Trifonov, V.A.; Giovannotti, M.; O’Brien, P.C.; Wallduck, M.; Lovell, F.; Rens, W.; Parise-Maltempi, P.P.; Caputo, V.; Ferguson-Smith, M.A. Chromosomal evolution in Gekkonidae. I. Chromosome painting between Gekko and Hemidactylus species reveals phylogenetic relationships within the group. Chromosome Res. 2011, 19, 843–855. [Google Scholar] [CrossRef]
- King, M. Chromosomal evolution in the Diplodactylinae (Gekkonidae: Reptila). I. Evolutionary relationships and patterns of change. Aust. J. Zool. 1987, 35, 507–531. [Google Scholar] [CrossRef]
- Oliver, P.; Hutchinson, M.; Hutchinson, R. Karyotypic Variation in the Australian Gecko Diplodactylus tessellatus, with the Description of a New Karyotypic Complement for Diplodactyline Geckos. J. Herpetol. 2007, 41, 540–543. [Google Scholar] [CrossRef]
- Amor, D.J.; Bentley, K.; Ryan, J.; Perry, J.; Wong, L.; Slater, H.; Choo, K.H. Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. USA 2004, 101, 6542–6547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, M.; Rofe, R. Karyotypic variation in the Australian Gekko Phyllodactylus marmoratus (Gray) (Gekkonidae: Reptilia). Chromosoma 1976, 54, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Aprea, G.; Andreone, F.; Fulgione, D.; Petraccioli, A.; Odierna, G. Chromosomal rearrangements occurred repeatedly and independently during species diversification in Malagasy geckos, genus Paroedura. Afr. Zool. 2013, 48, 96–108. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Aprea, G.; Glaw, F.; Odierna, G.; Guarino, F.M. Molecular phylogeny, biogeography and chromosome evolution of Malagasy dwarf geckos of the genus Lygodactylus (Squamata, Gekkonidae). Zool. Scr. 2017, 46, 42–54. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Guarino, F.; Loader, S.; Odierna, G.; Streicher, J.; Cooper, N. First karyological analysis of the endemic Malagasy phantom gecko Matoatoa brevipes (Squamata: Gekkonidae). Acta Herpetol. 2020, 15, 137–141. [Google Scholar]
- Gamble, T.A. Review of sex determining mechanisms in geckos (Gekkota: Squamata). Sex Dev. 2010, 4, 88–103. [Google Scholar] [CrossRef] [Green Version]
- Pokorná, M.; Rábová, M.; Ráb, P.; Ferguson-Smith, M.A.; Rens, W.; Kratochvíl, L. Differentiation of sex chromosomes and karyotypic evolution in the eye-lid geckos (Squamata: Gekkota: Eublepharidae), a group with different modes of sex determination. Chromosome Res. 2010, 18, 809–820. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Visone, V.; Petraccioli, A.; Odierna, G.; Capriglione, T.; Guarino, F.M. Non-random accumulation of LINE1-like sequences on differentiated snake W chromosomes. J. Zool. 2016, 300, 67–75. [Google Scholar] [CrossRef]

| Species | Specimen | Locality | Sex |
|---|---|---|---|
| B. antongilensis | GA 483 | Masobe Forest, Betampona | female |
| B. antongilensis | GA 1049 | Masobe Forest, Betampona | female |
| B. boivini | FGMV 3010 | Montagne des Français | female |
| B. sakalava clade A | FGMV 2029 | Ifaty | female |
| B. sakalava clade A | FGMV 2030 | Ifaty | male |
| B. sakalava clade A | FGMV 2032 | Ifaty | male |
| B. sakalava clade B | GA 80 | Isalo | male |
| B. sakalava clade B | GA 376 | Marofandilia | juvenile |
| B. sakalava clade B | GA 377 | Marofandilia | male |
| B. sakalava clade B | GA 378 | Marofandilia | male |
| Species | B. antongilensis | B. boivini | B. sakalava cl. A | B. sakalava cl.B |
|---|---|---|---|---|
| Chrom. | RL% | RL% | RL% | RL% |
| 1 | 9.6 ± 0.7 25.0 ± 4.0 (st/sm) | 9.7 ± 0.6 (t) | 9.8 ± 1.0 39.3 ± 4.5 (m) | 9.1 ± 0.8 43.1 ± 3.4 (m) |
| 2 | 8.7 ± 1.1 (t) | 8.2 ± 0.4 (t) | 8.2 ± 0.9 (t) | 8.1 ± 0.9 (t) |
| 3 | 7.4 ± 0.4 (t) | 7.4 ± 1.2 (t) | 8.1 ± 0.7 (t) | 7.8 ± 0.6 (t) |
| 4 | 7.3 ± 0.5 (t) | 6.6 ± 0.9 (t) | 7.1 ± 0.6 (t) | 6.6± 0.8 (t) |
| 5 | 7.0 ± 0.7 (t) | 5.8 ± 0.7 (t) | 6.8.1 ± 0.5 (t) | 6.4 ± 0.6 (t) |
| 6 | 6.7 ± 0.6 38.4 ± 3.5 (m) | 5.8 ± 1.0 (t) | 6.2 ± 0.8 47.0 ± 3.0 (m) | 6.2 ± 0.6 46.3 ± 3.1 (m) |
| 7 | 6.2 ± 0.9 (t) | 5.7 ± 0.5 (t) | 5.4 ± 0.6 (t) | 6.1 ± 0.8 (t) |
| 8 | 5.6 ± 0.8 (t) | 5.6 ± 0.4 (t) | 5.3 ± 0.7 (t) | 5.7 ± 0.6 (t) |
| 9 | 5.0 ± 0.6 (t) | 5.6 ± 0.6 (t) | 5.3 ±0.8 (t) | 5.4 ± 0.5 (t) |
| 10 | 4.9 ± 0.5 (t) | 5.6 ± 0.6 (t) | 5.2 ± 0,6 (t) | 4.9 ± 0.4 (t) |
| 11 | 4.8 ± 0.8 (t) | 4.6 ± 0.5 (t) | 4.2 ± 0.6 (t) | 4.5 ± 0.6 (t) |
| 12 | 4.6 ± 0.4 (t) | 3.9 ± 0.5 (t) | 4.0 ± 0.4 (t) | 4.1 ± 0.7 (t) |
| 13 | 4.0 ± 0.6 (t) | 3.3 ± 0.4 (t) | 3.8 ± 0.5 (t) | 3.8 ± 0.7 (t) |
| 14 | 3.3 ± 1.0 (t) | 3.3 ± 0.8 (t) | 3.7 ± 0.7 (t) | 3.6 ± 0.9 (t) |
| 15 | 3.2 ± 0.8 (t) | 3.2 ± 0.6 (t) | 3.7 ± 0.7 (t) | 3.4 ± 0.5 (t) |
| 16 | 2.8 ± 0.9 (t) | 3.2 ± 0.7 (t) | 3.5 ± 0.6 (t) | 3.1 ± 0.8 (t) |
| 17 | 2.6 ± 0.7 (t) | 2.9 ± 0.5 (t) | 3.3 ± 0.6 (t) | 3.0 ± 0.5 (t) |
| 18 | 2.3 ± 0.6 (t) | 2.8 ± 0.4 (t) | 2.8 ± 0.7 (t) | 2.8 ± 0.4 (t) |
| 19 | 2.0 ± 0.5 (t) | 2.4 ± 0.4 (t) | 2.7 ± 0.8 (t) | 2.7 ± 0.6 (t) |
| 20 | 2.0 ± 0.8 (t) | 2.3 ± 0.3 (t) | 2.6 ± 0.5 (t) | 2.6 ± 0.7 (t) |
| 21 | 1.9 ± 0.5 (t) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mezzasalma, M.; Brunelli, E.; Odierna, G.; Guarino, F.M. Chromosome Diversity and Evolution of the Endemic Malagasy Velvet Geckos of the Genus Blaesodactylus (Reptilia, Gekkonidae). Animals 2023, 13, 2068. https://doi.org/10.3390/ani13132068
Mezzasalma M, Brunelli E, Odierna G, Guarino FM. Chromosome Diversity and Evolution of the Endemic Malagasy Velvet Geckos of the Genus Blaesodactylus (Reptilia, Gekkonidae). Animals. 2023; 13(13):2068. https://doi.org/10.3390/ani13132068
Chicago/Turabian StyleMezzasalma, Marcello, Elvira Brunelli, Gaetano Odierna, and Fabio Maria Guarino. 2023. "Chromosome Diversity and Evolution of the Endemic Malagasy Velvet Geckos of the Genus Blaesodactylus (Reptilia, Gekkonidae)" Animals 13, no. 13: 2068. https://doi.org/10.3390/ani13132068






