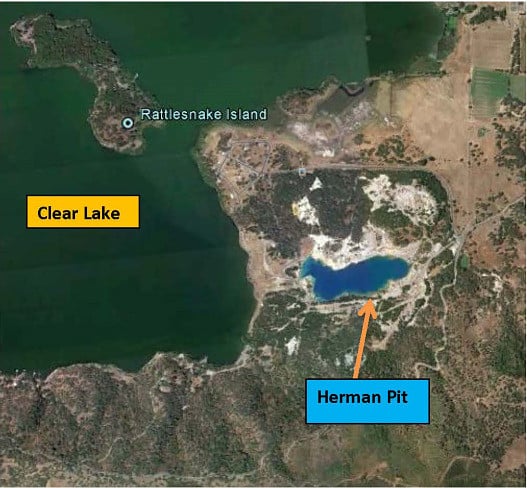
Methane Oxidation and Molecular Characterization of Methanotrophs from a Former Mercury Mine Impoundment
Abstract
:1. Introduction
2. Methods and Materials
2.1. Sediment Slurry Incubations
2.2. Enrichment Cultures
2.3. Analytical
2.4. Lipid Analysis
2.5. Phylogenetic Analysis
3. Results
3.1. Gases
| Component | Concentration | δ13C (‰) |
|---|---|---|
| CO2 | 95% | −11 |
| CH4 | 5% | −24 |
| C2H6 | 10–20 ppm | BD |
| C1/C2 + C3 | >5000 | ND |
| Dissolved CH4 | 400 μM | ND |
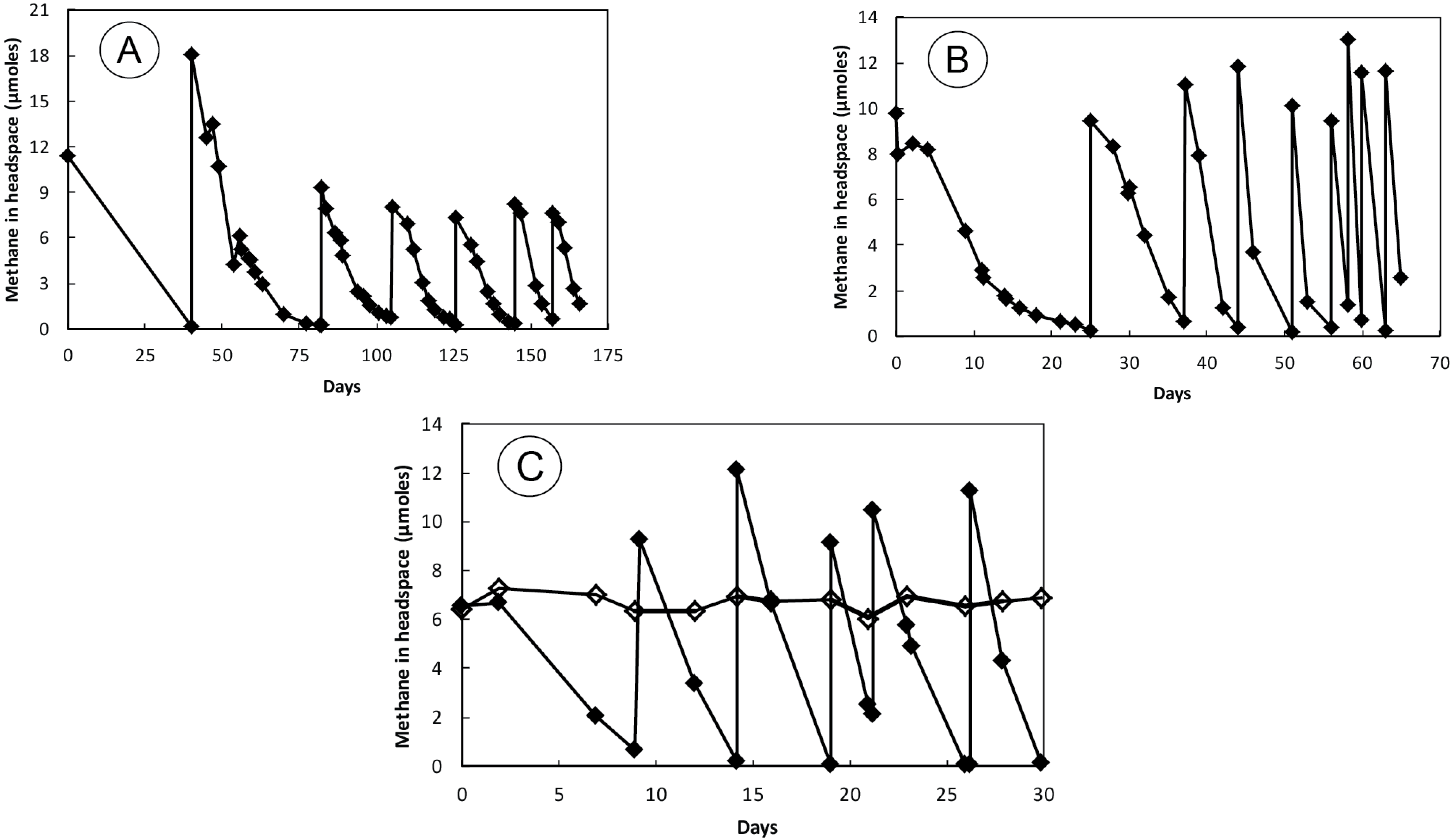
3.2. Sediment Incubations
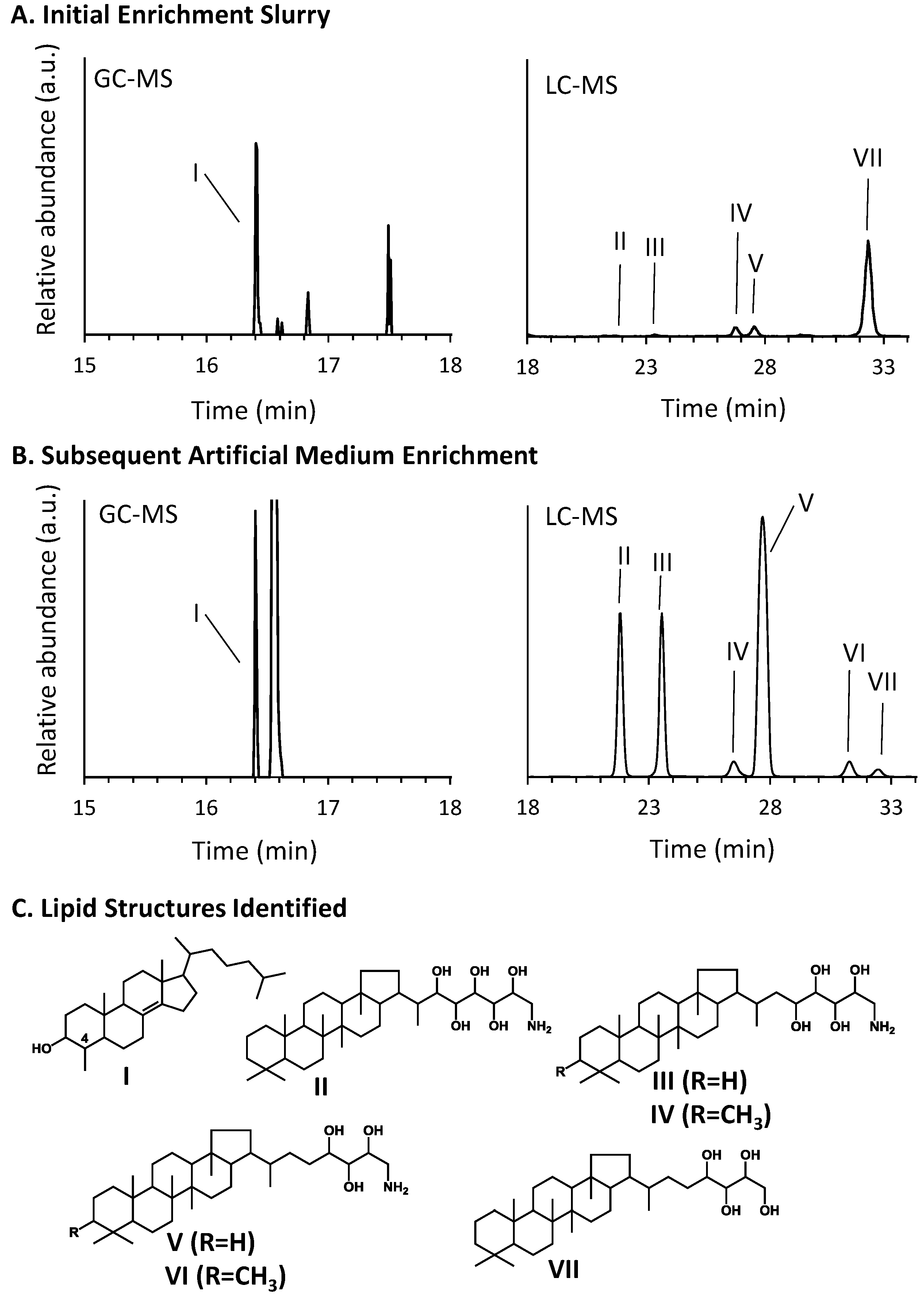
3.3. Hopanoid and Sterol Production
| Hopanoid Produced | μg hopanoid/μg total lipid extract | |
|---|---|---|
| Initial Slurry | Enrichment Culture | |
| V. Aminotriol | 0.2 | 82 |
| III. Aminotetrol | 0.04 | 30 |
| II. Aminopentol | 0.03 | 32 |
| VI. 3-Methylaminotriol | BD | 4 |
| IV. 3-Methylaminotetrol | 0.2 | 4 |
| VII. Bacteriohopanetetrol | 2 | 1 |
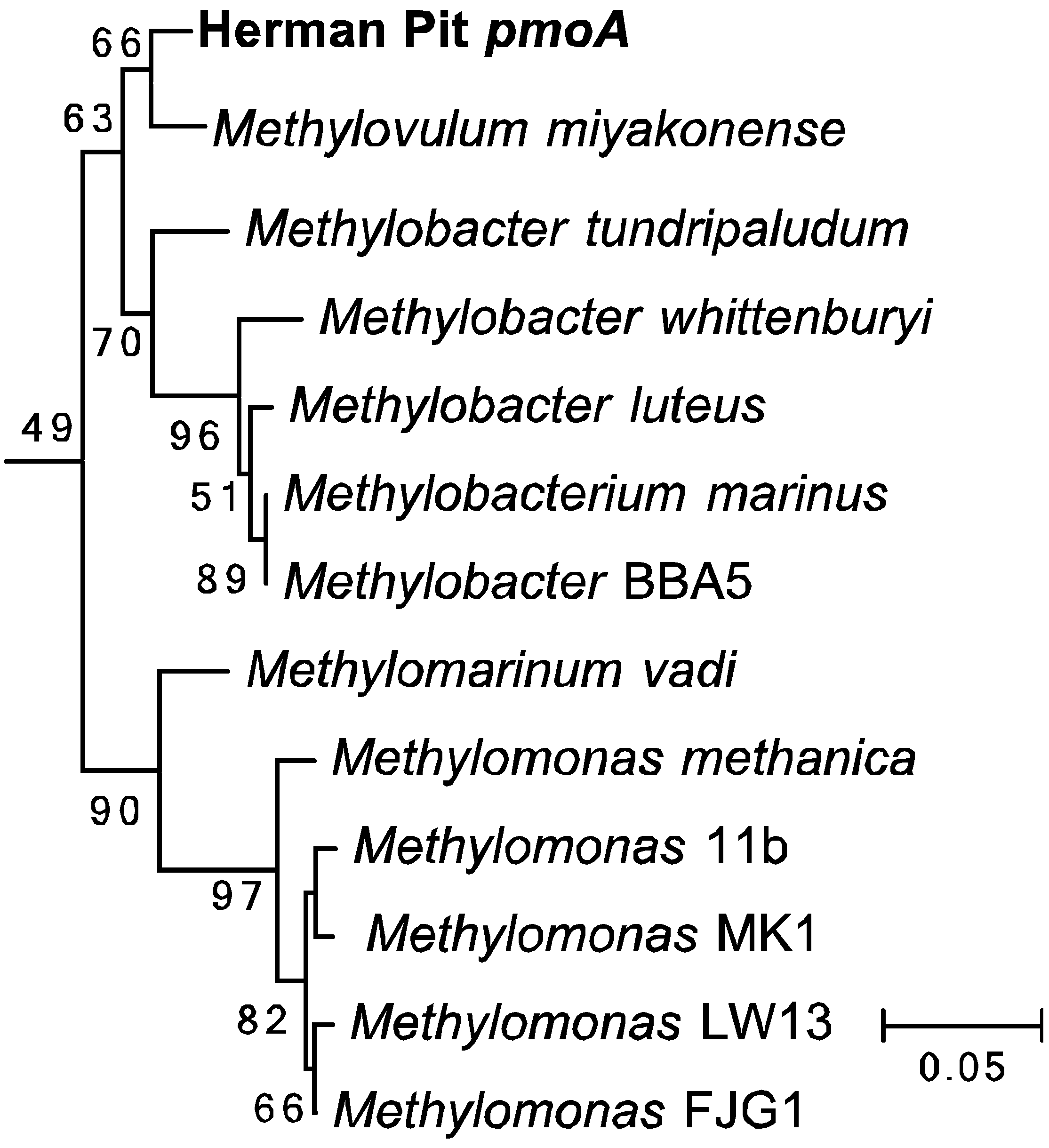
3.4. Amplification and Sequencing of 16S rRNA and pmoA Genes

3.5. Amplification and Sequencing of 16S rRNA and pmoA Genes

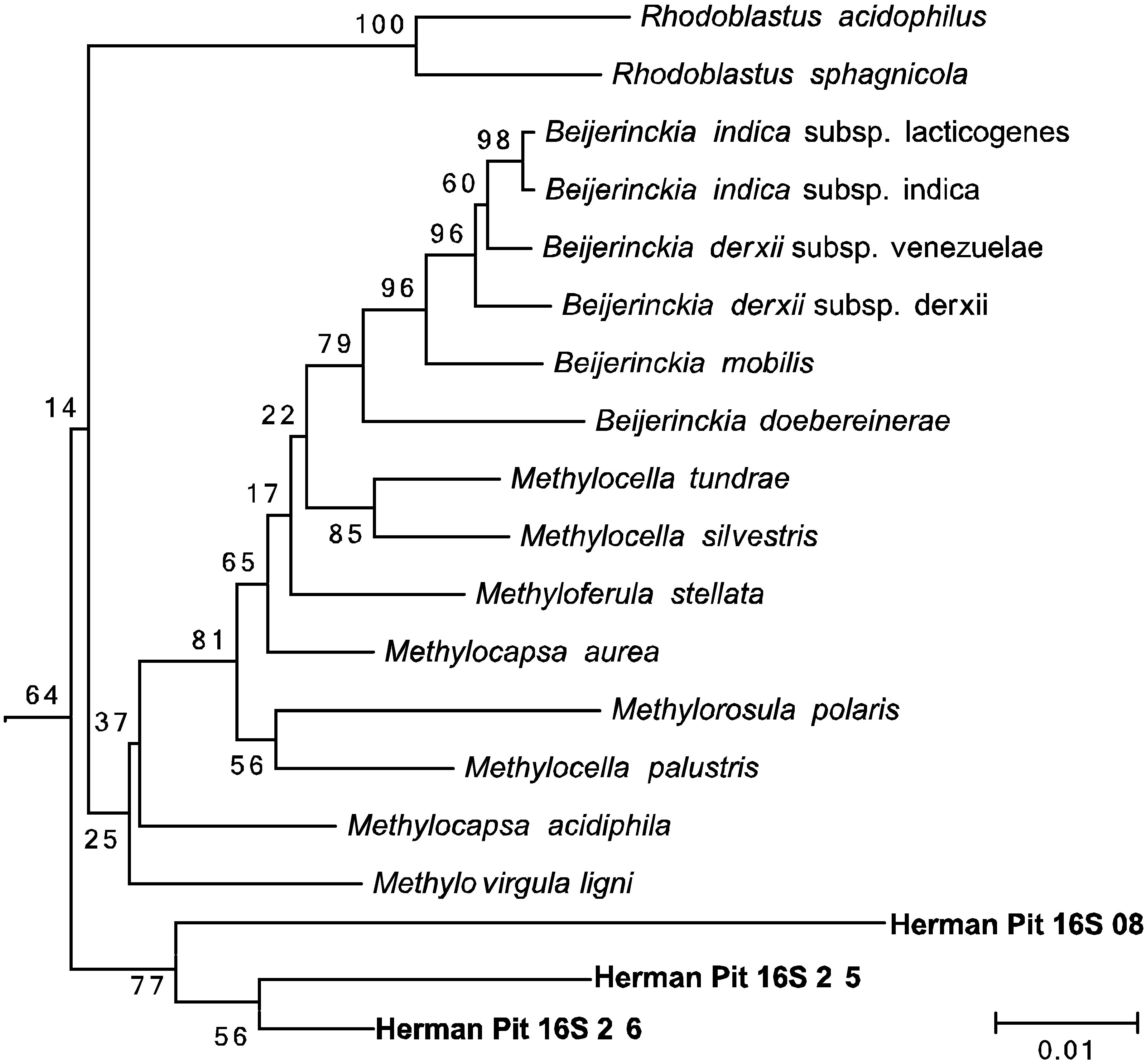
3.6. Enrichment Cultures
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Walter, K.M.; Zimov, S.A.; Chanton, J.P.; Verblya, D.; Chapin, F.S., III. Methane bubbling from Siberian thaw lakes as a positive feedback to climate warming. Nature 2006, 443, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Cicerone, R.; Oremland, R.S. Biogeochemical aspects of atmospheric methane. Glob. Biogeochem. Cycles 1988, 2, 299–328. [Google Scholar] [CrossRef]
- Reeburgh, W.S. Global methane biogeochemistry. In Treatise on Geochemistry; Keeling, R.F., Ed.; Elsevier: Amsterdam, The Netherlands, 2003; Volume 4. [Google Scholar]
- Ribbons, D.W.; Harrison, J.E.; Wadzinski, A.M. Metabolism of single carbon compounds. Annu. Rev. Microbiol. 1970, 24, 135–158. [Google Scholar] [CrossRef] [PubMed]
- Quayle, J.R. The metabolism of one-carbon compounds by micro-organisms. Adv. Microb. Physiol. 1972, 7, 119–203. [Google Scholar]
- Anthony, C. The Biochemistry of Methylotrophs; Academic Press: London, UK, 1982. [Google Scholar]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [PubMed]
- Söhngen, N.L. Ueber Bakterien, welche Methan als Kohlenstoffnahrung und Energiequelle gebrauchen. Centralbl. Bakteriol. Parasitenk. Infectionskr. Hyg. Abt. II 1906, 15, 513–517. [Google Scholar]
- Whittenbury, R.; Phillips, K.C.; Wilkinson, J.F. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 1970, 61, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Whittenbury, R.; Davies, S.L.; Davey, J.F. Exospores and cysts formed by methane-utilizing bacteria. J. Gen. Microbiol. 1970, 61, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.L.; Whittenbury, R. Fine structure of methane and other hydrocarbon-utilizing bacteria. J. Gen. Microbiol. 1970, 61, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Theisen, A.R.; Murrell, J.C. Facultative methanotrophs revisited. J. Bacteriol. 2005, 24, 4303–4305. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, P.F.; Kmelenina, V.N.; Suzina, N.E.; Trotsenko, Y.A.; Dedysh, S.N. Methylocella silvestris sp. nov., a novel methanotrophic bacterium isolated from an acidic forest cambisol. Int. J. Syst. Evol. Microbiol. 2003, 53, 1231–1239. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Dunfield, P.F.; Trotsenko, Y.A. Methane utilization by Methylobacterium species: New evidence but still no proof for an old controversy. Int. J. Syst. Evol. Microbiol. 2004, 54, 1919–1920. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Knief, C.; Dunfield, P.F. Methylocella species are facultatively methanotrophic. J. Bacteriol. 2005, 187, 4665–4670. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, P.F.; Yuryev, A.; Senin, P.; Smirova, A.; Stott, M.B.; Hou, S.; Ly, B.; Saw, J.H.; Zhou, Z.; Ren, Y.; et al. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 2007, 450, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Pol, A.; Heijmans, K.; Harhangi, H.R.; Tedesco, D.; Jetten, M.S.M.; Op den Camp, H.J.M. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 2007, 450, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Van Teeseling, M.C.F.; Pol, A.; Harhangi, H.R.; van der Zwart, S.; Jetten, M.S.M.; Op den Camp, H.J.M.; van Niftrik, L. Expanding the verrucomicrobial methanotrophic world: Description of three novel species of Methylacidimicrobium gen. nov. Appl. Environ. Microbiol. 2014, 80, 6792–6791. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, P.F. Methanotrophy in extreme environments. In Encyclopedia of Life Sciences; Wiley: Chichester, UK, 2009. [Google Scholar] [CrossRef]
- Engel, M.A.; Goff, F.; Jewett, D.G.; Reller, G.J.; Bauman, J.B. Application of environmental groundwater tracers at the Sulfur Bank mercury mine, California, USA. Hydrolgeol. J. 2008, 16, 559–573. [Google Scholar] [CrossRef]
- Wells, J.T.; Ghioso, M.S. Rock alteration, mercury transport, and metal deposition at Sulfur Bank, California. Econ. Geol. 1988, 83, 606–618. [Google Scholar] [CrossRef]
- Donnelly-Nolan, J.M.; Burns, M.G.; Goff, F.E.; Peters, E.K.; Thompson, J.M. The Geysers-Clear Lake Area, California: Thermal waters, mineralization, volcanism, and geothermal potential. Econ. Geol. 1993, 88, 301–316. [Google Scholar] [CrossRef]
- Nehring, N.L. Gases from springs and wells in the Geysers-Clear Lake Area; USGS Professional Paper 1141; U.S. Geological Survey: Washington, DC, USA, 1981; pp. 205–209.
- Lucero, J. USEPA Sulphur Bank Remedial Investigation and Feasibility Study. Unpublished work. 2013. [Google Scholar]
- Drummond, S.E.; Ohmoto, H. Chemical evolution and mineral deposition in boiling hydrothermal systems. Econ. Geol. 1985, 80, 126–147. [Google Scholar] [CrossRef]
- Kizilova, A.K.; Voryanchikova, E.; Sukhacheva, M.; Kravchenko, I.; Gal’chenko, V. Investigation of the methanotrophic communities of the hot springs of the Uzon caldera, Kamchatka, by molecular ecological techniques. Microbiology 2012, 81, 606–613. [Google Scholar] [CrossRef]
- Kizilova, A.K.; Sukhacheva, M.V.; Pimenov, N.V.; Yurkov, A.M.; Kravchenko, I.K. Methane oxidation activity and diversity of aerobic methanotrophs in pH-neutral and semi-neutral thermal springs of the Kinashir Island, Russian Far East. Extremophiles 2014, 18, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Silverman, M.P.; Lundgren, D.G. Studies on the chemoautotrophic iron bacterium Ferrobacillus ferrooxidans. J. Bacteriol. 1959, 77, 642–647. [Google Scholar] [PubMed]
- Oremland, R.S.; DesMarais, D.J. Distribution, abundance, and carbon isotopic composition of gaseous hydrocarbons in Big Soda Lake, Nevada: An alkaline, meromictic lake. Geochim. Cosmochim. Acta 1983, 47, 2107–2114. [Google Scholar] [CrossRef]
- Miller, L.G.; Oremland, R.S. Methane efflux from the pelagic regions of four lakes. Glob. Biogeochem. Cycles 1988, 3, 269–278. [Google Scholar] [CrossRef]
- Miller, L.G.; Baesman, S.M.; Kirshtein, J.; Voytek, M.A.; Oremland, R.S. A biogeochemical and genetic survey of acetylene fermentation by environmental samples and bacterial isolates. Geomicrobiol. J. 2013, 30, 501–516. [Google Scholar] [CrossRef]
- Oremland, R.S. Hydrogen metabolism by decomposing cyanobacterial aggregates in Big Soda Lake, Nevada. Appl. Environ. Microbiol. 1983, 45, 1519–1525. [Google Scholar] [PubMed]
- Crosson, E.R.; Ricci, K.N.; Richman, B.A.; Chilese, F.C.; Owano, T.G.; Provencal, R.A.; Todd, M.W.; Glasser, J.; Kachanov, A.A.; Paldus, B.A.; et al. Stable isotope ratios using cavity ring-down spectroscopy: Determination of 13C/12C for carbon dioxide in human breath. Anal. Chem. 2002, 74, 2003–2007. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Winderlich, J.; Gerbig, C.; Hoefer, A.; Crosson, E.R.; van Pelt, A.D.; Steinbach, J.; Kolle, O.; Beck, V.; Daube, B.C.; et al. High-energy continuous airborne measurements of greenhouse gases (CO2 and CH4) using the cavity ring-down spectroscopy (CRDS) technique. Atmos. Meas. Tech. 2010, 3, 375–386. [Google Scholar] [CrossRef]
- Welander, P.V.; Summons, R.E. Discovery, taxonomic distribution, and phenotypic characterization of a gene required for 3-methylhopanoid production. Proc. Natl. Acad. Sci. USA 2012, 109, 12905–12910. [Google Scholar] [CrossRef] [PubMed]
- Welander, P.V.; Hunter, R.C.; Zhang, L.; Sessions, A.L.; Summons, R.E.; Newman, D.K. Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 2009, 191, 6145–6156. [Google Scholar] [CrossRef] [PubMed]
- Welander, P.V.; Doughty, D.M.; Wu, C.H.; Mehay, S.; Summons, R.E.; Newman, D.K. Identification and characterization of Rhodopseudomonas palustris TIE-1 hopanoid biosynthesis mutants. Geobiology 2012, 10, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Talbot, H.M.; Watson, D.F.; Murrell, J.C.; Carter, J.F.; Farrimond, P. Analysis of intact bacteriohopanepolyols from methanotrophic bacteria by reversed-phase high-performance liquid chromatography-atmospheric pressure chemical ionisation mass spectrometry. J. Chromatogr. A 2001, 921, 175–185. [Google Scholar] [CrossRef]
- Talbot, H.M.; Summons, R.; Jahnke, L.; Farrimond, P. Characteristic fragmentation of bacteriohopanepolyols during atmospheric pressure chemical ionisation liquid chromatography/ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2788–2796. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.J.; Costello, A.; Lidstrom, M.E.; Murrell, J.C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 1995, 132, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Eden, P.A.; Schmidt, T.M.; Blakemore, R.P.; Pace, N.R. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 1991, 41, 324–325. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.G.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Francey, R.J.; Allison, C.E.; Etheridge, D.M.; Trudinger, C.M.; Enting, I.G.; Luenberger, M.; Lagensfeld, R.L.; Michel, E.; Steele, L.P. A 1000-year high precision record of δ13C in atmospheric CO2. Tellus 1999, 51B, 170–193. [Google Scholar] [CrossRef]
- Longinelli, A.; Lenaz, R.; Ori, C.; Selmo, E. Concentrations and δ13C values of atmospheric CO2 from oceanic atmosphere through time: Polluted and non-polluted areas. Tellus 2005, 57, 385–390. [Google Scholar] [CrossRef]
- Gleason, J.D.; Kyser, T.K. Stable isotope compositions of gases and vegetation near naturally burning coal. Nature 1984, 307, 254–257. [Google Scholar] [CrossRef]
- Whiticar, M.J.; Faber, E.; Schoell, M. Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation-isotope evidence. Geochim. Cosmochim. Acta 1986, 50, 693–709. [Google Scholar] [CrossRef]
- Oremland, R.S.; Miller, L.G.; Whiticar, M.J. Sources and flux of natural gas from Mono Lake, California. Geochim. Cosmochim. Acta 1987, 51, 2915–2929. [Google Scholar] [CrossRef]
- Bernard, B.B.; Brooks, J.M.; Sackett, W.M. Light hydrocarbons in recent Texas continental shelf and slope sediments. J. Geophys. Res. 1978, 83, 4053–4061. [Google Scholar] [CrossRef]
- Volkman, J.K. Sterols and other triterpenoids: Source specificity and evolution of biosynthetic pathways. Org. Geochem. 2005, 36, 139–159. [Google Scholar] [CrossRef]
- Talbot, H.M.; Handley, L.; Spencer-Jones, C.; Bienvenu, D.J.; Schefuß, E.; Mann, P.; Poulson, J.; Spencer, R.; Wagner, T. Variability in aerobic methane oxidation over the past 1.2 Myrs recorded in microbial biomarker signatures from Congo fan sediments. Geochim. Cosmochim. Acta 2014, 133, 387–401. [Google Scholar] [CrossRef]
- Lin, J.L.; Joye, S.B.; Scholten, J.C.; Schafer, H.; McDonald, I.R.; Murrell, J.C. Analysis of methane monooxygenase genes in Mono Lake suggests that increased methane oxidation activity may correlate with a change in methanotroph community structure. Appl. Environ. Microbiol. 2005, 7, 6458–6462. [Google Scholar] [CrossRef] [PubMed]
- Kip, N.; Ouyang, W.; van Winden, J.; Raghoebarsing, A.; van Niftrik, L.; Pol, A.; Pan, Y.; Bodrossy, L.; van Donselaar, E.G.; Reichart, G.J.; et al. Detection, isolation, and characterization of acidophilic methanotrophs from sphagnum mosses. Appl. Environ. Microbiol. 2011, 77, 5643–5654. [Google Scholar] [CrossRef] [PubMed]
- Weaver, T.L.; Dugan, P.R. Enhancement of bacterial methane oxidation by clay minerals. Nature 1972, 237, 518. [Google Scholar] [CrossRef] [PubMed]
- Rudd, J.W.M.; Hamilton, R.D.; Campbell, N.E.R. Measurement of microbial oxidation of methane in lake water. Limnol. Oceanogr. 1974, 19, 519–524. [Google Scholar] [CrossRef]
- Zaiss, U.; Winter, P.; Kaltwasser, H. Microbial methane oxidation in the Saar River. Z. Allg. Mikrobiol. 1982, 22, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Abramochkina, F.N.; Bezrukova, L.V.; Koshelev, A.V.; Gal’chenko, V.F.; Ivanov, M.V. Microbial oxidation of methane in a body of fresh water. Microbiologia 1987, 56, 464–471. [Google Scholar]
- Remsen, C.C.; Minnich, E.C.; Stephens, R.S.; Buchholz, L.; Lidstrom, M.E. Methane oxidation in Lake Superior sediments. J. Great Lakes Res. 1989, 15, 141–146. [Google Scholar] [CrossRef]
- Frenzel, P.; Thebrath, B.; Conrad, R. Oxidation of methane in the oxic surface layer of a deep lake sediment (Lake Constance). FEMS Microbiol. Ecol. 1990, 73, 149–158. [Google Scholar] [CrossRef]
- Pol, A.; Barends, T.R.M.; Dietl, A.; Khadem, A.F.; Eygensteyn, J.; Jetten, M.S.M.; Op den Camp, H.J.M. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 2014, 16, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, H.; Yurimoto, H.; Sakai, Y. Stimulation of methanotrophic growth in cocultures by cobalamin excreted by rhizobia. Appl. Environ. Microbiol. 2011, 77, 8509–8515. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.; de Roy, K.; Thas, O.; de Neve, J.; Hoefman, S.; Vandamme, P.; Heylen, K.; Boon, N. The more the merrier: Heterotroph richness stimulates methanotrophic activity. ISME J. 2014, 8, 1945–1948. [Google Scholar] [CrossRef] [PubMed]
- Oremland, R.S.; Culberston, C.W.; Winfrey, M.R. Methylmercury decomposition in sediments and bacterial cultures: Involvement of methanogens and sulfate reducers in oxidative demethylation. Appl. Environ. Microbiol. 1991, 57, 130–137. [Google Scholar] [PubMed]
- Oremland, R.S.; Miller, L.G.; Dowdle, P.; Connell, T.; Barkay, T. Methylmercury oxidative demethylation potentials in contaminated and pristine sediments of the Carson River, Nevada. Appl. Environ. Microbiol. 1995, 61, 2745–2753. [Google Scholar] [PubMed]
- Marvin-DiPasquale, M.C.; Oremland, R.S. Bacterial methylmercury degradation in Florida everglades peat sediment. Environ. Sci. Technol. 1998, 32, 2556–2563. [Google Scholar] [CrossRef]
- Marvin-DiPasquale, M.C.; Agree, J.; McGowan, C.; Oremland, R.S.; Thomas, M.; Krabbenhoft, D.; Gilmour, C.C. Methyl-mercury degradation pathways: A comparison among three mercury-impacted ecosystems. Environ. Sci. Technol. 2000, 34, 4908–4916. [Google Scholar] [CrossRef]
- Boden, R.; Murrell, C. Response to mercury (II) ions in Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 2011, 324, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Vorobev, A.; Jagadevan, S.; Baral, B.S.; DiSpirito, A.A.; Freemeir, B.C.; Bergman, B.H.; Bandow, N.L.; Semrau, J.D. Detoxification of mercury by methanobactin from Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2013, 79, 5918–5926. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baesman, S.M.; Miller, L.G.; Wei, J.H.; Cho, Y.; Matys, E.D.; Summons, R.E.; Welander, P.V.; Oremland, R.S. Methane Oxidation and Molecular Characterization of Methanotrophs from a Former Mercury Mine Impoundment. Microorganisms 2015, 3, 290-309. https://doi.org/10.3390/microorganisms3020290
Baesman SM, Miller LG, Wei JH, Cho Y, Matys ED, Summons RE, Welander PV, Oremland RS. Methane Oxidation and Molecular Characterization of Methanotrophs from a Former Mercury Mine Impoundment. Microorganisms. 2015; 3(2):290-309. https://doi.org/10.3390/microorganisms3020290
Chicago/Turabian StyleBaesman, Shaun M., Laurence G. Miller, Jeremy H. Wei, Yirang Cho, Emily D. Matys, Roger E. Summons, Paula V. Welander, and Ronald S. Oremland. 2015. "Methane Oxidation and Molecular Characterization of Methanotrophs from a Former Mercury Mine Impoundment" Microorganisms 3, no. 2: 290-309. https://doi.org/10.3390/microorganisms3020290
APA StyleBaesman, S. M., Miller, L. G., Wei, J. H., Cho, Y., Matys, E. D., Summons, R. E., Welander, P. V., & Oremland, R. S. (2015). Methane Oxidation and Molecular Characterization of Methanotrophs from a Former Mercury Mine Impoundment. Microorganisms, 3(2), 290-309. https://doi.org/10.3390/microorganisms3020290