Transcription and Maturation of mRNA in Dinoflagellates
Abstract
:1. Introduction
2. Transcription and Its Regulation
2.1. cis-Acting Sequences and RNA Polymerase Components
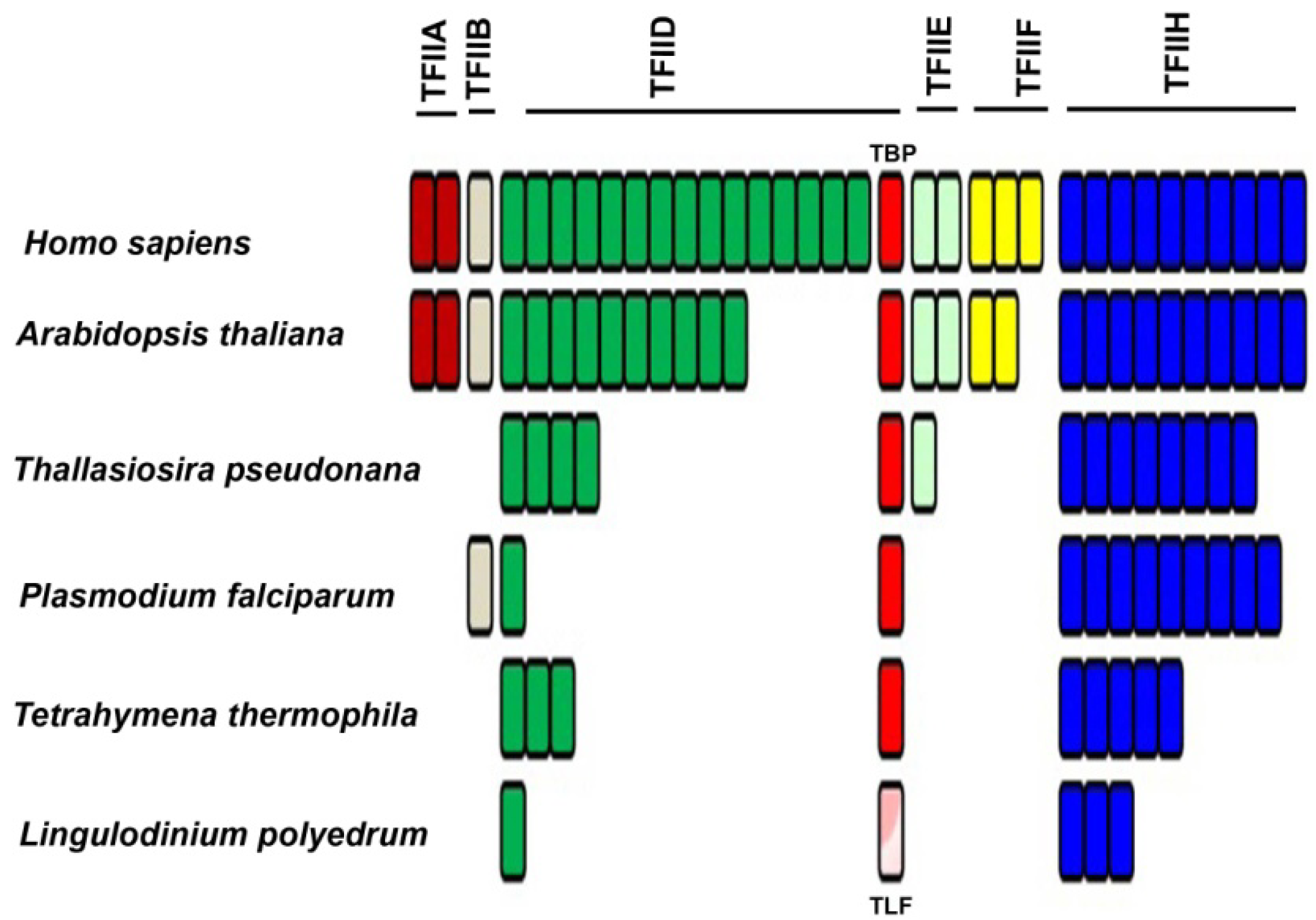
2.2. Basal/General Transcription Factors
2.3. DNA Binding Proteins
2.4. Transcriptional Regulation
3. Splicing and the Spliceosome
4. RNA Transport and mRNA Surveillance Pathways
| Mammal | Plant | Alveolata | Diatom | ||||
|---|---|---|---|---|---|---|---|
| Lp | Pf | Tt | |||||
| Nucleus | 11 | 10 | 6 | 9 | 7 | 8 | |
| Central channel | Nuclear basket | 4 | 1 | 1 | 0 | 0 | 1 |
| Symmetrical nups | 11 | 9 | 2 | 1 | 4 | 6 | |
| Central channel | 3 | 3 | 0 | 0 | 0 | 1 | |
| Spoke complex | 5 | 5 | 0 | 0 | 0 | 2 | |
| Lumenal ring | 3 | 1 | 0 | 0 | 1 | 0 | |
| Cytoplasmic tails | 8 | 6 | 2 | 2 | 3 | 3 | |
| Cytoplasm | 53 | 37 | 17 | 17 | 17 | 24 | |
| Mammal | Plant | Alveolata | Diatom | ||||
|---|---|---|---|---|---|---|---|
| Lp | Pf | Tt | |||||
| Nucleus | Cap binding complex | 2 | 2 | 0 | 1 | 2 | 1 |
| EJC | 15 | 11 | 5 | 4 | 4 | 5 | |
| 5′ capping | 2 | 2 | 0 | 0 | 1 | 2 | |
| Pre-mRNA processing | 14 | 13 | 4 | 4 | 4 | 8 | |
| Cytoplasm | Nonsense mediated decay | 12 | 9 | 7 | 6 | 5 | 6 |
| No-go decay | 3 | 3 | 3 | 2 | 2 | 3 | |
5. Conclusions and Perspectives
Supplementary Materials
Supplementary File 1Acknowledgments
Conflicts of Interest
References
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef]
- Muscatine, L.; McCloskey, L.R.; Marian, R.E. Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnol. Oceanogr. 1981, 26, 601–611. [Google Scholar] [CrossRef]
- Camacho, F.G.; Rodriguez, J.G.; Miron, A.S.; Garcia, M.C.; Belarbi, E.H.; Chisti, Y.; Grima, E.M. Biotechnological significance of toxic marine dinoflagellates. Biotechnol. Adv. 2007, 25, 176–194. [Google Scholar] [CrossRef]
- Schmitter, R.E.; Njus, D.; Sulzman, F.M.; Gooch, V.D.; Hastings, J.W. Dinoflagellate bioluminescence: A comparative study of in vitro components. J. Cell. Physiol. 1976, 87, 123–134. [Google Scholar] [CrossRef]
- Hastings, J.W.; Sweeney, B.M. A persistent diurnal rhythm of luminescence in Gonyaulax polyedra. Biol. Bull. 1958, 115, 444–458. [Google Scholar]
- Hastings, J.W.; Astrachan, L.; Sweeney, B.M. A persistent daily rhythm in photosynthesis. J. Gen. Physiol. 1961, 45, 69–76. [Google Scholar] [CrossRef]
- Sweeney, B.M. The photosynthetic rhythm in single cells of Gonyaulax polyedra. Cold Spring Harb. Symp. Quant. Biol. 1960, 25, 145–148. [Google Scholar] [CrossRef]
- Roenneberg, T.; Colfax, G.N.; Hastings, J.W. A circadian rhythm of population behavior in Gonyaulax polyedra. J. Biol. Rhythms 1989, 4, 201–216. [Google Scholar]
- Hastings, J.W. The Gonyaulax clock at 50: Translational control of circadian expression. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 141–144. [Google Scholar] [CrossRef]
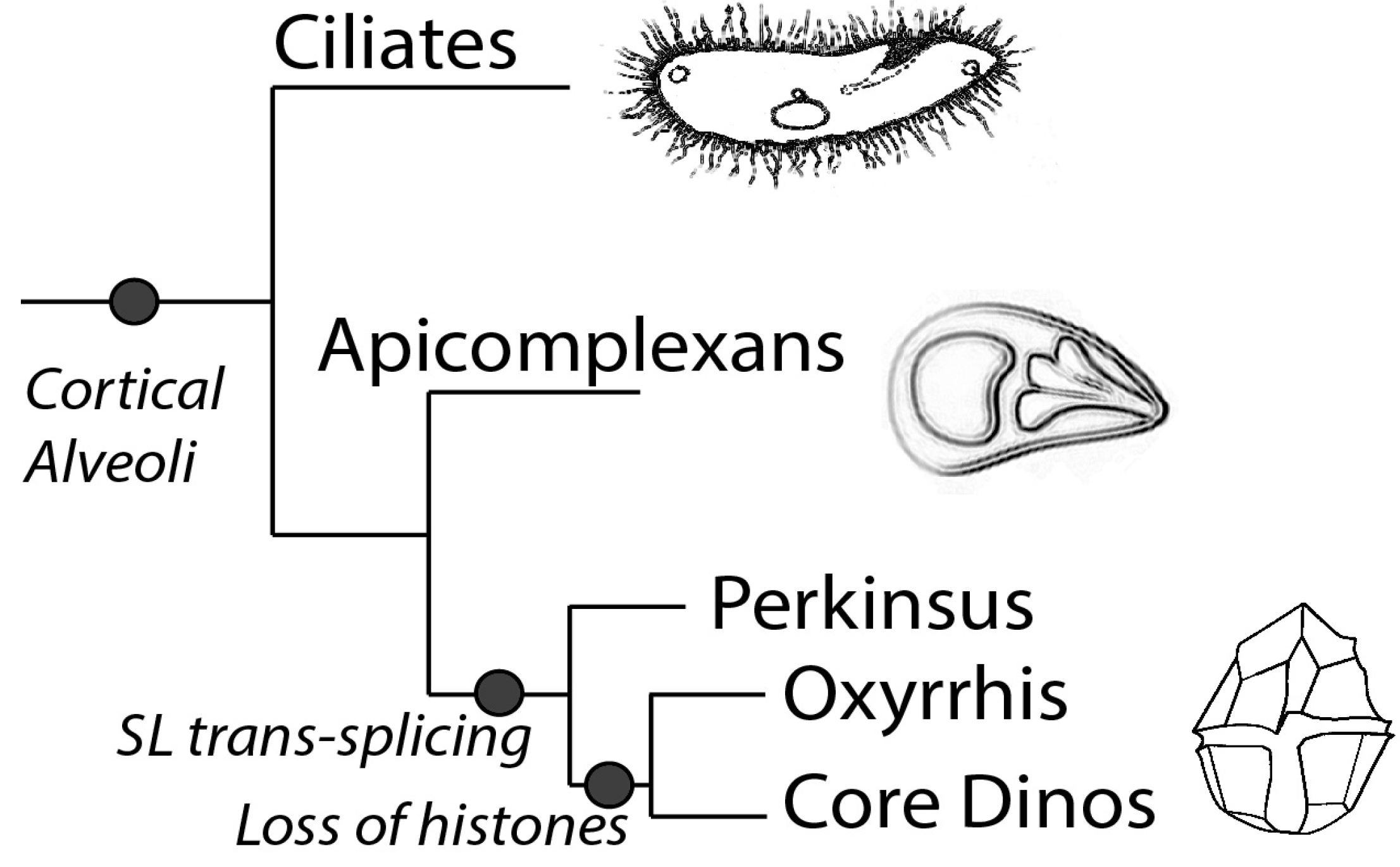
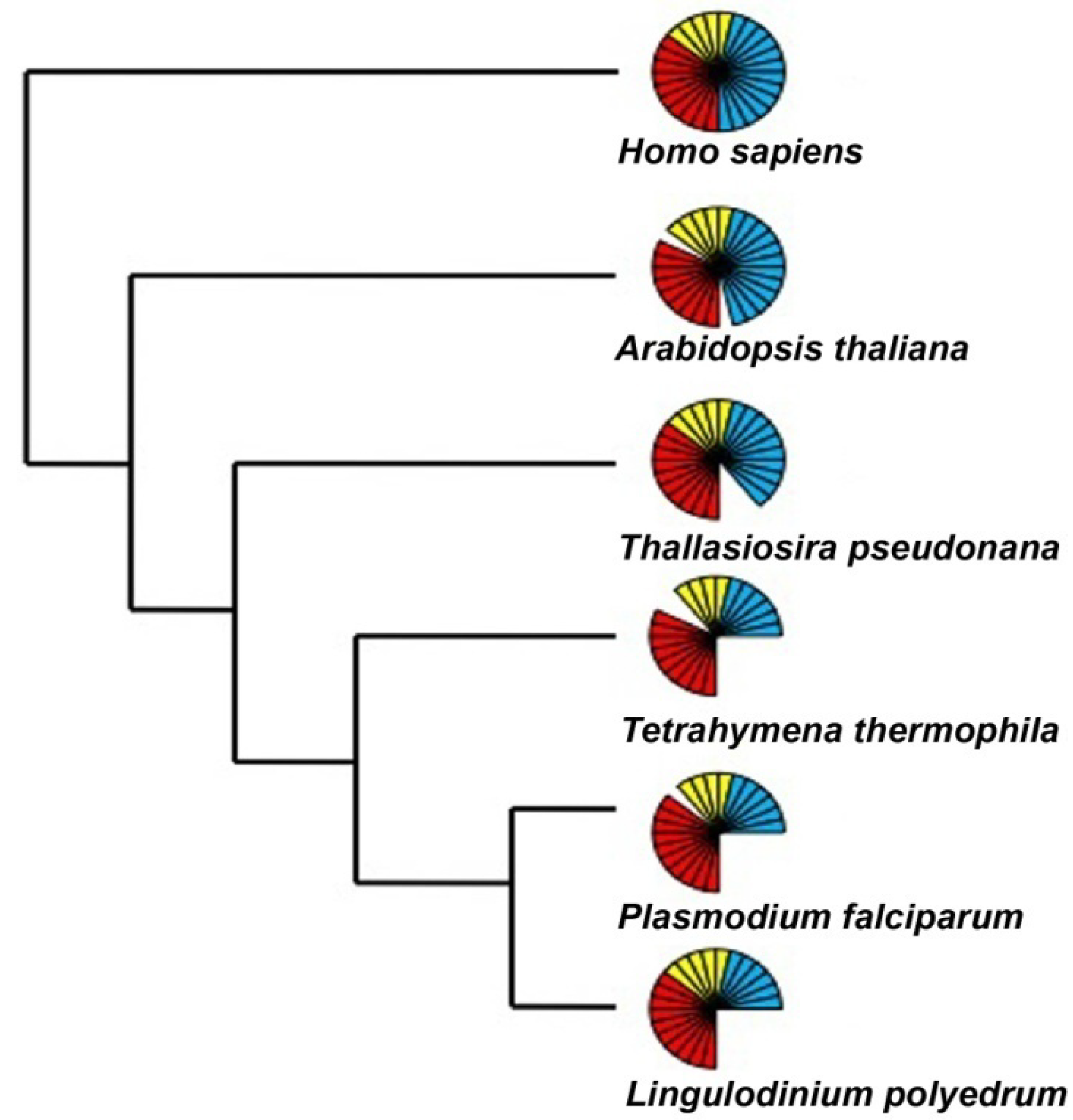
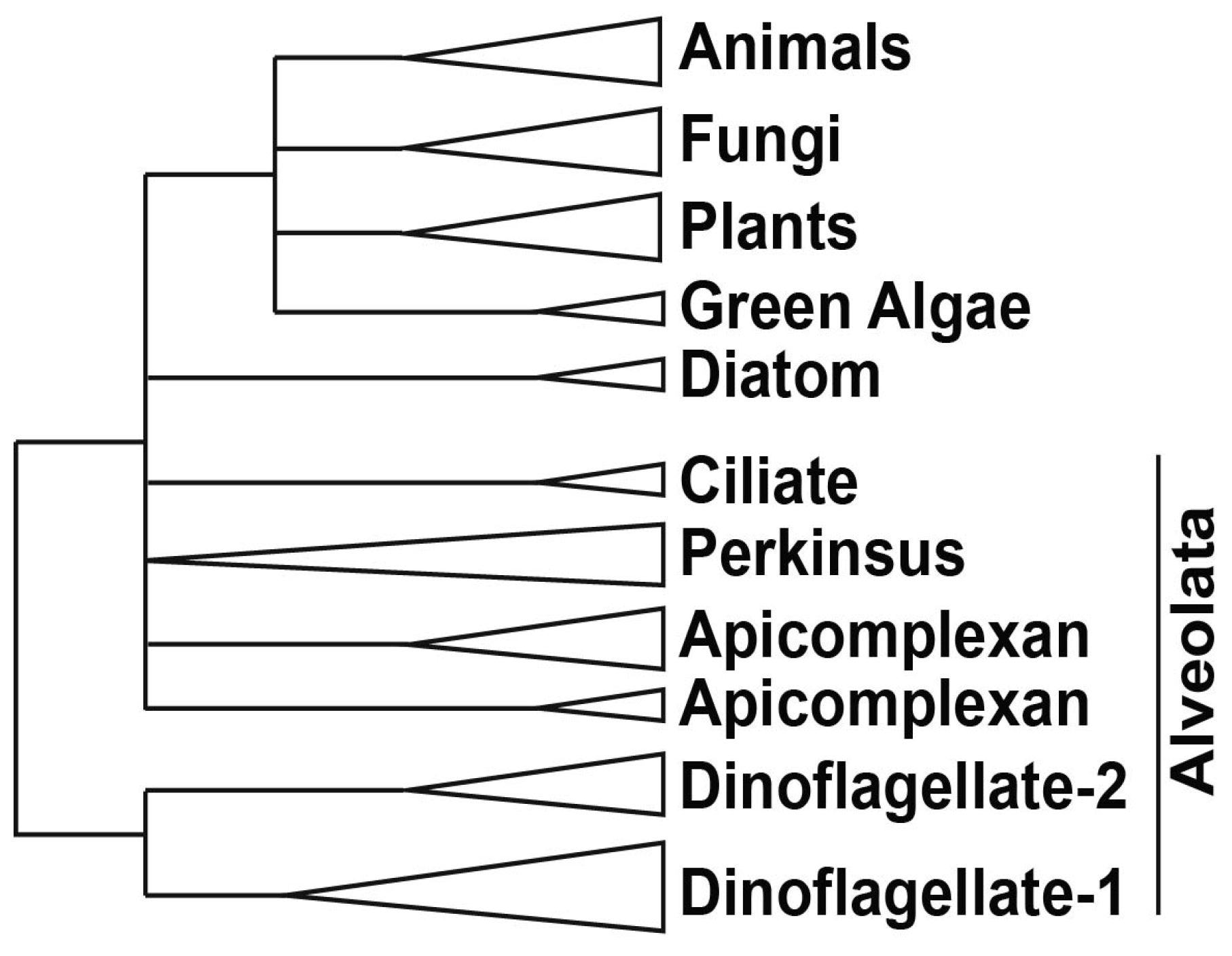
- Fast, N.M.; Xue, L.; Bingham, S.; Keeling, P.J. Re-examining alveolate evolution using multiple protein molecular phylogenies. J. Eukaryot. Microbiol. 2002, 49, 30–37. [Google Scholar] [CrossRef]
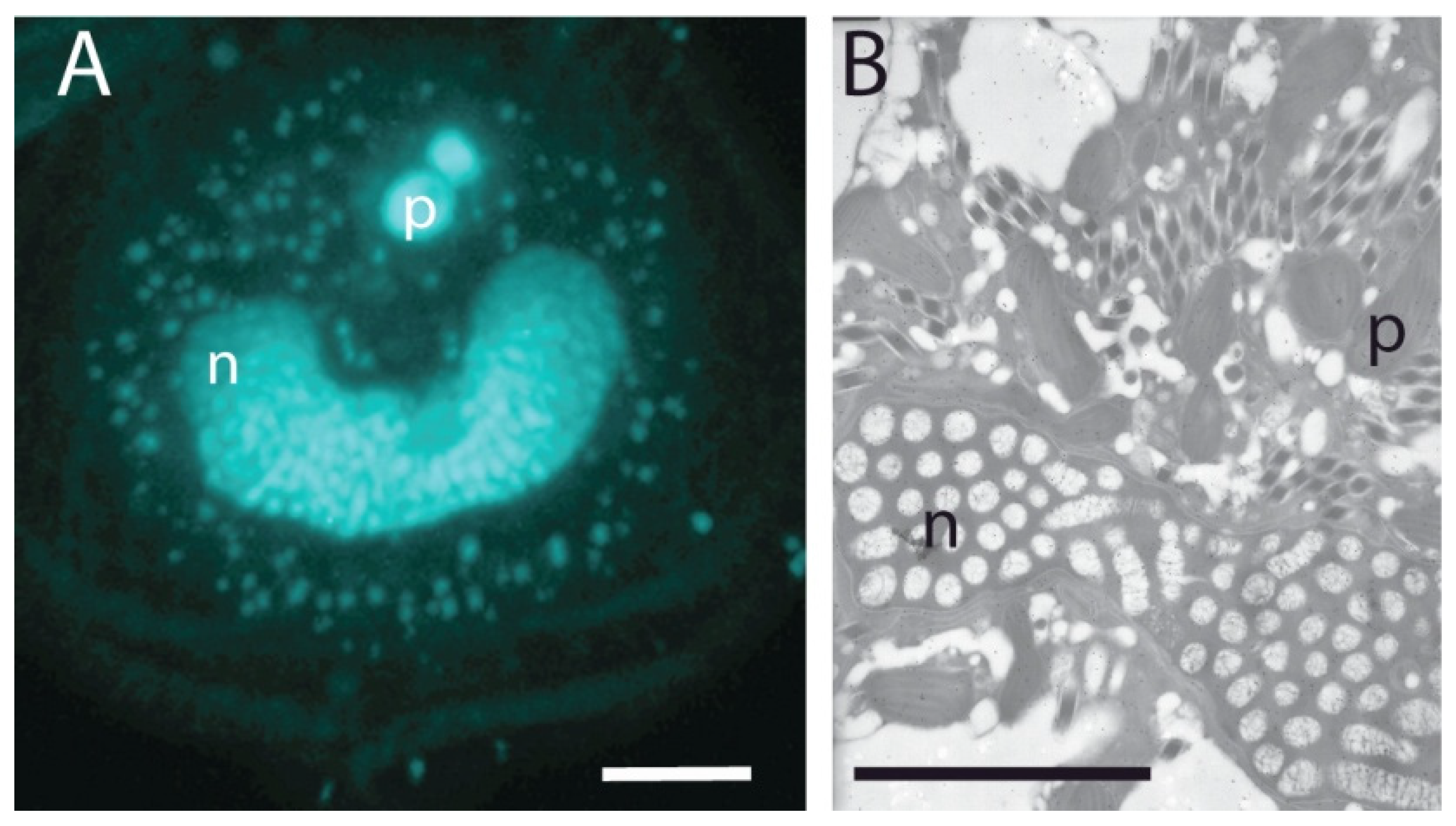
- Spector, D.L. Dinoflagellate Nuclei. In Dinoflagellates; Spector, D.L., Ed.; Academic Press: London, UK, 1984; pp. 107–147. [Google Scholar]
- Lin, S. Genomic understanding of dinoflagellates. Res. Microbiol. 2011, 162, 551–569. [Google Scholar] [CrossRef]
- Wisecaver, J.H.; Hackett, J.D. Dinoflagellate genome evolution. Annu. Rev. Microbiol. 2011, 65, 369–387. [Google Scholar] [CrossRef]
- Hackett, J.D.; Anderson, D.M.; Erdner, D.L.; Bhattacharya, D. Dinoflagellates: A remarkable evolutionary experiment. Am. J. Bot. 2004, 91, 1523–1534. [Google Scholar]
- Livolant, F. Cholesteric organization of DNA in vivo and in vitro. Eur. J. Cell Biol. 1984, 33, 300–311. [Google Scholar]
- Livolant, F. Positive and negative birefringence in chromosomes. Chromosoma 1978, 68, 45–58. [Google Scholar] [CrossRef]
- Herzog, M.; Soyer, M.O. The native structure of dinoflagellate chromosomes and their stabilization by Ca2+ and Mg2+ cations. Eur. J. Cell Biol. 1983, 30, 33–41. [Google Scholar]
- Sigee, D.C. Structural DNA and genetically active DNA in dinoflagellate chromosomes. Biosystems 1983, 16, 203–210. [Google Scholar]
- Kornberg, R.D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 12955–12961. [Google Scholar]
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef]
- Hahn, S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004, 11, 394–403. [Google Scholar]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.A.; Taylor, M.S.; Engstrom, P.G.; Frith, M.C.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar]
- Everett, R.D.; Baty, D.; Chambon, P. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983, 11, 2447–2464. [Google Scholar]
- Yoshikawa, T.; Takishita, K.; Ishida, Y.; Uchida, A. Molecular cloning and nucleotide sequence analysis of the gene coding for chloroplast-type ferredoxin from the dinoflagellates Peridinium bipes and Alexandrium tamarense. Fish. Sci. 1997, 63, 692–700. [Google Scholar]
- Wong, J.M.; Liu, F.; Bateman, E. Isolation of genomic DNA encoding transcription factor TFIID from Acanthamoeba castellanii: Characterization of the promoter. Nucleic Acids Res. 1992, 20, 4817–4824. [Google Scholar] [CrossRef]
- Huang, W.; Bateman, E. Cloning, expression, and characterization of the TATA-binding protein (TBP) promoter binding factor, a transcription activator of the Acanthamoeba TBP gene. J. Biol. Chem. 1995, 270, 28839–28847. [Google Scholar] [CrossRef]
- Cohen, S.M.; Knecht, D.; Lodish, H.F.; Loomis, W.F. DNA sequences required for expression of a Dictyostelium actin gene. EMBO J. 1986, 5, 3361–3366. [Google Scholar]
- Kimmel, A.R.; Firtel, R.A. Sequence organization in Dictyostelium: Unique structure at the 5′-ends of protein coding genes. Nucleic Acids Res. 1983, 11, 541–552. [Google Scholar] [CrossRef]
- Liston, D.R.; Johnson, P.J. Analysis of a ubiquitous promoter element in a primitive eukaryote: Early evolution of the initiator element. Mol. Cell. Biol. 1999, 19, 2380–2388. [Google Scholar]
- McAndrew, M.B.; Read, M.; Sims, P.F.; Hyde, J.E. Characterisation of the gene encoding an unusually divergent TATA-binding protein (TBP) from the extremely A+T-rich human malaria parasite Plasmodium falciparum. Gene 1993, 124, 165–171. [Google Scholar]
- Luo, H.; Gilinger, G.; Mukherjee, D.; Bellofatto, V. Transcription initiation at the TATA-less spliced leader RNA gene promoter requires at least two DNA-binding proteins and a tripartite architecture that includes an initiator element. J. Biol. Chem. 1999, 274, 31947–31954. [Google Scholar]
- Quon, D.V.; Delgadillo, M.G.; Johnson, P.J. Transcription in the early diverging eukaryote Trichomonas vaginalis: An unusual RNA polymerase II and alpha-amanitin-resistant transcription of protein-coding genes. J. Mol. Evol. 1996, 43, 253–262. [Google Scholar]
- Quon, D.V.; Delgadillo, M.G.; Khachi, A.; Smale, S.T.; Johnson, P.J. Similarity between a ubiquitous promoter element in an ancient eukaryote and mammalian initiator elements. Proc. Natl. Acad. Sci. USA 1994, 91, 4579–4583. [Google Scholar] [CrossRef]
- Le, Q.H.; Markovic, P.; Hastings, J.W.; Jovine, R.V.; Morse, D. Structure and organization of the peridinin-chlorophyll a-binding protein gene in Gonyaulax polyedra. Mol. Gen. Genet. 1997, 255, 595–604. [Google Scholar]
- Li, L.; Hastings, J.W. The structure and organization of the luciferase gene in the photosynthetic dinoflagellate Gonyaulax polyedra. Plant Mol. Biol. 1998, 36, 275–284. [Google Scholar] [CrossRef]
- Machabee, S.; Wall, L.; Morse, D. Expression and genomic organization of a dinoflagellate gene family. Plant Mol. Biol. 1994, 25, 23–31. [Google Scholar] [CrossRef]
- Lee, D.H.; Mittag, M.; Sczekan, S.; Morse, D.; Hastings, J.W. Molecular cloning and genomic organization of a gene for luciferin-binding protein from the dinoflagellate Gonyaulax polyedra. J. Biol. Chem. 1993, 268, 8842–8850. [Google Scholar]
- Okamoto, O.K.; Liu, L.; Robertson, D.L.; Hastings, J.W. Members of a dinoflagellate luciferase gene family differ in synonymous substitution rates. Biochemistry 2001, 40, 15862–15868. [Google Scholar] [CrossRef]
- Bachvaroff, T.R.; Place, A.R. From stop to start: Tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS One 2008, 3, e2929. [Google Scholar] [CrossRef]
- Jackson, A.P. Tandem gene arrays in Trypanosoma brucei: Comparative phylogenomic analysis of duplicate sequence variation. BMC Evol. Biol. 2007, 7. [Google Scholar] [CrossRef]
- Beauchemin, M.; Roy, S.; Daoust, P.; Dagenais-Bellefeuille, S.; Bertomeu, T.; Letourneau, L.; Lang, B.F.; Morse, D. Dinoflagellate tandem array gene transcripts are highly conserved and not polycistronic. Proc. Natl. Acad. Sci. USA 2012, 109, 15793–15798. [Google Scholar] [CrossRef]
- Rizzo, P.J. RNA synthesis in isolated nuclei of the dinoflagellate Crypthecodinium cohnii. J. Protozool. 1979, 26, 290–294. [Google Scholar]
- Palenchar, J.B.; Bellofatto, V. Gene transcription in trypanosomes. Mol. Biochem. Parasitol. 2006, 146, 135–141. [Google Scholar] [CrossRef]
- Orphanides, G.; Lagrange, T.; Reinberg, D. The general transcription factors of RNA polymerase II. Genes Dev. 1996, 10, 2657–2683. [Google Scholar] [CrossRef]
- Conaway, J.W.; Bond, M.W.; Conaway, R.C. An RNA polymerase II transcription system from rat liver. Purification of an essential component. J. Biol. Chem. 1987, 262, 8293–8297. [Google Scholar]
- Conaway, R.C.; Conaway, J.W. An RNA polymerase II transcription factor has an associated DNA-dependent ATPase (dATPase) activity strongly stimulated by the TATA region of promoters. Proc. Natl. Acad. Sci. USA 1989, 86, 7356–7360. [Google Scholar] [CrossRef]
- Conaway, J.W.; Conaway, R.C. A multisubunit transcription factor essential for accurate initiation by RNA polymerase II. J. Biol. Chem. 1989, 264, 2357–2362. [Google Scholar]
- Conaway, J.W.; Reines, D.; Conaway, R.C. Transcription initiated by RNA polymerase II and purified transcription factors from liver. Cooperative action of transcription factors τ and ε in initial complex formation. J. Biol. Chem. 1990, 265, 7552–7558. [Google Scholar]
- Sumimoto, H.; Ohkuma, Y.; Yamamoto, T.; Horikoshi, M.; Roeder, R.G. Factors involved in specific transcription by mammalian RNA polymerase II: Identification of general transcription factor TFIIG. Proc. Natl. Acad. Sci. USA 1990, 87, 9158–9162. [Google Scholar] [CrossRef]
- Poon, D.; Bai, Y.; Campbell, A.M.; Bjorklund, S.; Kim, Y.J.; Zhou, S.; Kornberg, R.D.; Weil, P.A. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1995, 92, 8224–8228. [Google Scholar]
- Sanders, S.L.; Weil, P.A. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 2000, 275, 13895–13900. [Google Scholar] [CrossRef]
- Chatterjee, S.; Struhl, K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature 1995, 374, 820–822. [Google Scholar] [CrossRef]
- Chong, J.A.; Moran, M.M.; Teichmann, M.; Kaczmarek, J.S.; Roeder, R.; Clapham, D.E. TATA-binding protein (TBP)-like factor (TLF) is a functional regulator of transcription: Reciprocal regulation of the neurofibromatosis type 1 and c-fos genes by TLF/TRF2 and TBP. Mol. Cell. Biol. 2005, 25, 2632–2643. [Google Scholar]
- Holmes, M.C.; Tjian, R. Promoter-selective properties of the TBP-related factor TRF1. Science 2000, 288, 867–870. [Google Scholar] [CrossRef]
- Guillebault, D.; Sasorith, S.; Derelle, E.; Wurtz, J.M.; Lozano, J.C.; Bingham, S.; Tora, L.; Moreau, H. A new class of transcription initiation factors, intermediate between TATA box-binding proteins (TBPs) and TBP-like factors (TLFs), is present in the marine unicellular organism, the dinoflagellate Crypthecodinium cohnii. J. Biol. Chem. 2002, 277, 40881–40886. [Google Scholar] [CrossRef]
- Bayer, T.; Aranda, M.; Sunagawa, S.; Yum, L.K.; Desalvo, M.K.; Lindquist, E.; Coffroth, M.A.; Voolstra, C.R.; Medina, M. Symbiodinium transcriptomes: Genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One 2012, 7, e35269. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Toulza, E.; Shin, M.S.; Blanc, G.; Audic, S.; Laabir, M.; Collos, Y.; Claverie, J.M.; Grzebyk, D. Gene expression in proliferating cells of the dinoflagellate Alexandrium catenella (Dinophyceae). Appl. Environ. Microbiol. 2010, 76, 4521–4529. [Google Scholar] [CrossRef]
- Qiu, X.B.; Lin, Y.L.; Thome, K.C.; Pian, P.; Schlegel, B.P.; Weremowicz, S.; Parvin, J.D.; Dutta, A. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem. 1998, 273, 27786–27793. [Google Scholar]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Eickbush, T.H.; Moudrianakis, E.N. The histone core complex: An octamer assembled by two sets of protein-protein interactions. Biochemistry 1978, 17, 4955–4964. [Google Scholar] [CrossRef]
- Kasinsky, H.E.; Lewis, J.D.; Dacks, J.B.; Ausio, J. Origin of H1 linker histones. FASEB J. 2001, 15, 34–42. [Google Scholar] [CrossRef]
- Rizzo, P.J. Those amazing dinoflagellate chromosomes. Cell Res. 2003, 13, 215–217. [Google Scholar] [CrossRef]
- Rizzo, P.J. Comparative aspects of basic chromatin proteins in dinoflagellates. Biosystems 1981, 14, 433–443. [Google Scholar]
- Vernet, G.; Sala-Rovira, M.; Maeder, M.; Jacques, F.; Herzog, M. Basic nuclear proteins of the histone-less eukaryote Crypthecodinium cohnii (Pyrrhophyta): Two-dimensional electrophoresis and DNA-binding properties. Biochim. Biophys. Acta 1990, 1048, 281–289. [Google Scholar] [CrossRef]
- Bodansky, S.; Mintz, L.B.; Holmes, D.S. The mesokaryote Gyrodinium cohnii lacks nucleosomes. Biochem. Biophys. Res. Commun. 1979, 88, 1329–1336. [Google Scholar] [CrossRef]
- Rizzo, P.J.; Nooden, L.D. Chromosomal proteins in the dinoflagellate alga Gyrodinium cohnii. Science 1972, 176, 796–797. [Google Scholar]
- Livolant, F. Cholesteric organization of DNA in the stallion sperm head. Tissue Cell 1984, 16, 535–555. [Google Scholar] [CrossRef]
- Balhorn, R. The protamine family of sperm nuclear proteins. Genome Biol. 2007, 8. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, H.; Zhuang, Y.; Tran, B.; Gill, J. Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc. Natl. Acad. Sci. USA 2010, 107, 20033–20038. [Google Scholar]
- Roy, S.; Morse, D. A full suite of histone and histone modifying genes are transcribed in the dinoflagellate Lingulodinium. PLoS One 2012, 7, e34340. [Google Scholar]
- Rizzo, P.J.; Jones, M.; Ray, S.M. Isolation and properties of isolated nuclei from the Florida red tide dinoflagellate Gymnodinium breve (Davis). J. Protozool. 1982, 29, 217–222. [Google Scholar] [CrossRef]
- Kellenberger, E.; Arnold-Schulz-Gahmen, B. Chromatins of low-protein content: Special features of their compaction and condensation. FEMS Microbiol. Lett. 1992, 79, 361–370. [Google Scholar]
- Holck, A.; Lossius, I.; Aasland, R.; Haarr, L.; Kleppe, K. DNA- and RNA-binding proteins of chromatin from Escherichia coli. Biochim. Biophys. Acta 1987, 908, 188–199. [Google Scholar] [CrossRef]
- Wong, J.T.; New, D.C.; Wong, J.C.; Hung, V.K. Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot. Cell 2003, 2, 646–650. [Google Scholar] [CrossRef]
- Sala-Rovira, M.; Geraud, M.L.; Caput, D.; Jacques, F.; Soyer-Gobillard, M.O.; Vernet, G.; Herzog, M. Molecular cloning and immunolocalization of two variants of the major basic nuclear protein (HCc) from the histone-less eukaryote Crypthecodinium cohnii (Pyrrhophyta). Chromosoma 1991, 100, 510–518. [Google Scholar] [CrossRef]
- Chudnovsky, Y.; Li, J.F.; Rizzo, P.J.; Hastings, J.W.; Fagan, T. Cloning, expression, and characterization of a histone-like protein from the marine dinoflagellate Lingulodinium polyedrum. J. Phycol. 2002, 38, 543–550. [Google Scholar]
- Gornik, S.G.; Ford, K.L.; Mulhern, T.D.; Bacic, A.; McFadden, G.I.; Waller, R.F. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr. Biol. 2012, 22, 2303–2312. [Google Scholar] [CrossRef]
- Azevedo, C. Fine structure of Perkinsus atlanticus n. sp. (Apicomplexa, Perkinsea) parasite of the clam Ruditapes decussatus from Portugal. J. Parasitol. 1989, 75, 627–635. [Google Scholar] [CrossRef]
- Jaeckisch, N.; Yang, I.; Wohlrab, S.; Glockner, G.; Kroymann, J.; Vogel, H.; Cembella, A.; John, U. Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii. PLoS One 2011, 6, e28012. [Google Scholar]
- Minguez, A.; Franca, S.; Moreno Diaz de la Espina, S. Dinoflagellates have a eukaryotic nuclear matrix with lamin-like proteins and topoisomerase II. J. Cell Sci. 1994, 107, 2861–2873. [Google Scholar]
- Dechat, T.; Pfleghaar, K.; Sengupta, K.; Shimi, T.; Shumaker, D.K.; Solimando, L.; Goldman, R.D. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008, 22, 832–853. [Google Scholar]
- Dechat, T.; Adam, S.A.; Goldman, R.D. Nuclear lamins and chromatin: When structure meets function. Adv. Enzym. Regul. 2009, 49, 157–166. [Google Scholar] [CrossRef]
- Dechat, T.; Adam, S.A.; Taimen, P.; Shimi, T.; Goldman, R.D. Nuclear lamins. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Bhaud, Y.; Geraud, M.L.; Ausseil, J.; Soyer-Gobillard, M.O.; Moreau, H. Cyclic expression of a nuclear protein in a dinoflagellate. J. Eukaryot. Microbiol. 1999, 46, 259–267. [Google Scholar] [CrossRef]
- Guillebault, D.; Derelle, E.; Bhaud, Y.; Moreau, H. Role of nuclear WW domains and proline-rich proteins in dinoflagellate transcription. Protist 2001, 152, 127–138. [Google Scholar]
- Boggon, T.J.; Shan, W.S.; Santagata, S.; Myers, S.C.; Shapiro, L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 1999, 286, 2119–2125. [Google Scholar] [CrossRef]
- Babu, M.M.; Luscombe, N.M.; Aravind, L.; Gerstein, M.; Teichmann, S.A. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004, 14, 283–291. [Google Scholar] [CrossRef]
- Sommerville, J. Activities of cold-shock domain proteins in translation control. Bioessays 1999, 21, 319–325. [Google Scholar]
- Balaji, S.; Babu, M.M.; Iyer, L.M.; Aravind, L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005, 33, 3994–4006. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Blank, R.J.; Huss, V.A.R.; Kersten, W. Base composition of DNA from symbiotic dinoflagellates: A tool for phylogenetic classification. Arch. Microbiol. 1988, 149, 515–520. [Google Scholar] [CrossRef]
- Steele, R.E.; Rae, P.M. Ordered distribution of modified bases in the DNA of a dinoflagellate. Nucleic Acids Res. 1980, 8, 4709–4725. [Google Scholar] [CrossRef]
- Ten Lohuis, M.R.; Miller, D.J. Light-regulated transcription of genes encoding peridinin chlorophyll a proteins and the major intrinsic light-harvesting complex proteins in the dinoflagellate amphidinium carterae hulburt (Dinophycae). Changes In cytosine methylation accompany photoadaptation. Plant Physiol. 1998, 117, 189–196. [Google Scholar] [CrossRef]
- Rae, P.M.; Steele, R.E. Modified bases in the DNAs of unicellular eukaryotes: An examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems 1978, 10, 37–53. [Google Scholar] [CrossRef]
- Teebor, G.W.; Frenkel, K.; Goldstein, M.S. Ionizing radiation and tritium transmutation both cause formation of 5-hydroxymethyl-2′-deoxyuridine in cellular DNA. Proc. Natl. Acad. Sci. USA 1984, 81, 318–321. [Google Scholar] [CrossRef]
- Rae, P.M. Hydroxymethyluracil in eukaryote DNA: A natural feature of the pyrrophyta (dinoflagellates). Science 1976, 194, 1062–1064. [Google Scholar]
- Thomas, S.; Green, A.; Sturm, N.R.; Campbell, D.A.; Myler, P.J. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics 2009, 10. [Google Scholar] [CrossRef]
- Wong, J.T.; Kwok, A.C. Proliferation of dinoflagellates: Blooming or bleaching. Bioessays 2005, 27, 730–740. [Google Scholar] [CrossRef]
- Gyula, P.; Schafer, E.; Nagy, F. Light perception and signalling in higher plants. Curr. Opin. Plant Biol. 2003, 6, 446–452. [Google Scholar] [CrossRef]
- Van Dolah, F.M.; Lidie, K.B.; More, J.S.; Brunelle, S.A.; Ryan, J.C.; Monroe, E.A.; Haynes, B.L. Microarray analysis of diurnal- and circadian-regulated genes in the Florida red-tide dinoflagellate Karenia brevis (Dinophyceae). J. Phycol. 2007, 43, 741–752. [Google Scholar] [CrossRef]
- Lesser, M.P. Elevated temperatures and ultraviolet radiation cause oxidative stress and inhibit photosynthesis in symbiotic dinoflagellates. Limnol. Oceanogr. 1996, 41, 271–283. [Google Scholar] [CrossRef]
- Lesser, M.P. Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs 1997, 16, 187–192. [Google Scholar]
- Rosic, N.N.; Pernice, M.; Dove, S.; Dunn, S.; Hoegh-Guldberg, O. Gene expression profiles of cytosolic heat shock proteins Hsp70 and Hsp90 from symbiotic dinoflagellates in response to thermal stress: Possible implications for coral bleaching. Cell Stress Chaperones 2010, 16, 69–80. [Google Scholar]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J., Jr. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 2005, 44, 7342–7372. [Google Scholar] [CrossRef]
- Okamoto, O.K.; Asano, C.S.; Aidar, E.; Colepicolo, P. Of cadmium on growth and superoxide dismutase activity of this species of dinoflagellate. The marine microalga Tetraselmis gracilis. J. Phycol. 1996, 32, 74–79. [Google Scholar]
- Okamoto, O.K.; Colepicolo, P. Response of superoxide dismutase to pollutant metal stress in the marine dinoflagellate Gonyaulax polyedra. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1998, 119, 67–73. [Google Scholar] [CrossRef]
- Okamoto, O.K.; Robertson, D.L.; Fagan, T.F.; Hastings, J.W.; Colepicolo, P. Different regulatory mechanisms modulate the expression of a dinoflagellate iron-superoxide dismutase. J. Biol. Chem. 2001, 276, 19989–19993. [Google Scholar]
- Okamoto, O.K.; Hastings, J.W. Genome-wide analysis of redox-regulated genes in a dinoflagellate. Gene 2003, 321, 73–81. [Google Scholar] [CrossRef]
- Guo, R.; Ebenezer, V.; Ki, J.S. Transcriptional responses of heat shock protein 70 (Hsp70) to thermal, bisphenol A, and copper stresses in the dinoflagellate Prorocentrum minimu. Chemosphere 2012, 89, 512–520. [Google Scholar] [CrossRef]
- Guo, R.; Ki, J.S. Differential transcription of heat shock protein 90 (HSP90) in the dinoflagellate Prorocentrum minimum by copper and endocrine-disrupting chemicals. Ecotoxicology 2012, 21, 1448–1457. [Google Scholar] [CrossRef]
- Lowe, C.D.; Mello, L.V.; Samatar, N.; Martin, L.E.; Montagnes, D.J.; Watts, P.C. The transcriptome of the novel dinoflagellate Oxyrrhis marina (Alveolata: Dinophyceae): Response to salinity examined by 454 sequencing. BMC Genomics 2011, 12. [Google Scholar] [CrossRef]
- Kondo, T.; Ishiura, M. The circadian clock of cyanobacteria. Bioessays 2000, 22, 10–15. [Google Scholar] [CrossRef]
- McClung, C.R. Plant circadian rhythms. Plant Cell 2006, 18, 792–803. [Google Scholar] [CrossRef]
- Loros, J.J.; Dunlap, J.C. Genetic and molecular analysis of circadian rhythms in Neurospora. Annu. Rev. Physiol. 2001, 63, 757–794. [Google Scholar] [CrossRef]
- Rivkees, S.A. The development of circadian rhythms: From animals to humans. Sleep Med. Clin. 2007, 2, 331–341. [Google Scholar] [CrossRef]
- Roenneberg, T.; Rehman, J. Nitrate, a nonphotic signal for the circadian system. FASEB J. 1996, 10, 1443–1447. [Google Scholar]
- Roenneberg, T.; Merrow, M. Entrainment of the human circadian clock. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 293–299. [Google Scholar] [CrossRef]
- Merrow, M.; Roenneberg, T. Circadian entrainment of Neurospora crassa. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 279–285. [Google Scholar] [CrossRef]
- Roenneberg, T.; Kumar, C.J.; Merrow, M. The human circadian clock entrains to sun time. Curr. Biol. 2007, 17, R44–R45. [Google Scholar] [CrossRef]
- Woelfle, M.A.; Johnson, C.H. No promoter left behind: Global circadian gene expression in cyanobacteria. J. Biol. Rhythms 2006, 21, 419–431. [Google Scholar] [CrossRef]
- Ito, H.; Mutsuda, M.; Murayama, Y.; Tomita, J.; Hosokawa, N.; Terauchi, K.; Sugita, C.; Sugita, M.; Kondo, T.; Iwasaki, H. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. USA 2009, 106, 14168–14173. [Google Scholar] [CrossRef]
- Okamoto, O.K.; Hastings, J.W. Novel dinoflagellate clock-related genes identified through microarray analysis. J. Phycol. 2003, 39, 519–526. [Google Scholar]
- Walz, B.; Walz, A.; Sweeney, B.M. A circadian rhythm in RNA in the dinoflagellate, Gonyaulax polyedra. J. Comp. Physiol. 1983, 151, 207–213. [Google Scholar]
- Dagenais-Bellefeuille, S.; Bertomeu, T.; Morse, D. S-phase and M-phase timing are under independent circadian control in the dinoflagellate Lingulodinium. J. Biol. Rhythms 2008, 23, 400–408. [Google Scholar] [CrossRef]
- Bertomeu, T.; Rivoal, J.; Morse, D. A dinoflagellate CDK5-like cyclin-dependent kinase. Biol. Cell 2007, 99, 531–540. [Google Scholar] [CrossRef]
- Karakashian, M.W.; Hastings, J.W. The inhibition of a biological clock by actinomycin D. Proc. Natl. Acad. Sci. USA 1962, 48, 2130–2137. [Google Scholar] [CrossRef]
- Rossini, C.; Taylor, W.; Fagan, T.; Hastings, J.W. Lifetimes of mRNAs for clock-regulated proteins in a dinoflagellate. Chronobiol. Int. 2003, 20, 963–976. [Google Scholar] [CrossRef]
- Morey, J.S.; Monroe, E.A.; Kinney, A.L.; Beal, M.; Johnson, J.G.; Hitchcock, G.L.; van Dolah, F.M. Transcriptomic response of the red tide dinoflagellate, Karenia brevis, to nitrogen and phosphorus depletion and addition. BMC Genomics 2011, 12. [Google Scholar] [CrossRef]
- Lin, X.; Zhang, H.; Huang, B.; Lin, S. Alkaline phosphatase gene sequence and transcriptional regulation by phosphate limitation in Amphidinium carterae (dinophyceae). J. Phycol. 2011, 47, 1110–1120. [Google Scholar] [CrossRef]
- Lin, X.; Zhang, H.; Huang, B.; Lin, S. Alkaline phosphatase gene sequence characteristics and transcriptional regulation by phosphate limitation in Karenia brevis (Dinophyceae). Harmful Algae 2012, 17, 14–24. [Google Scholar]
- Lee, T.C.; Kwok, O.T.; Ho, K.C.; Lee, F.W. Effects of different nitrate and phosphate concentrations on the growth and toxin production of an Alexandrium tamarense strain collected from Drake Passage. Mar. Environ. Res. 2012, 81, 62–69. [Google Scholar] [CrossRef]
- Yang, I.; Beszteri, S.; Tillmann, U.; Cembella, A.; John, U. Growth- and nutrient-dependent gene expression in the toxigenic marine dinoflagellate Alexandrium minutum. Harmful Algae 2011, 12, 55–69. [Google Scholar] [CrossRef] [Green Version]
- Moustafa, A.; Evans, A.N.; Kulis, D.M.; Hackett, J.D.; Erdner, D.L.; Anderson, D.M.; Bhattacharya, D. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS One 2010, 5, e9688. [Google Scholar] [CrossRef]
- Johnson, J.G.; Morey, J.S.; Neely, M.G.; Ryan, J.C.; van Dolah, F.M. Transcriptome remodeling associated with chronological aging in the dinoflagellate, Karenia brevis. Mar. Genomics 2012, 5, 15–25. [Google Scholar]
- Yang, I.; John, U.; Beszteri, S.; Glockner, G.; Krock, B.; Goesmann, A.; Cembella, A.D. Comparative gene expression in toxic versus non-toxic strains of the marine dinoflagellate Alexandrium minutum. BMC Genomics 2010, 11. [Google Scholar] [CrossRef]
- Salcedo, T.; Upadhyay, R.J.; Nagasaki, K.; Bhattacharya, D. Dozens of toxin-related genes are expressed in a nontoxic strain of the dinoflagellate Heterocapsa circularisquama. Mol. Biol. Evol. 2012, 29, 1503–1506. [Google Scholar] [CrossRef]
- Nassoury, N.; Cappadocia, M.; Morse, D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J. Cell Sci. 2003, 116, 2867–2874. [Google Scholar] [CrossRef]
- Shi, X.; Zhang, H.; Lin, S. Tandem repeats, high copy number and remarkable diel expression rhythm of form II RuBisCO in Prorocentrum donghaiense (dinophyceae). PLoS One 2013, 8, e71232. [Google Scholar]
- Gast, R.J.; Beaudoin, D.J.; Caron, D.A. Isolation of symbiotically expressed genes from the dinoflagellate symbiont of the solitary radiolarian Thalassicolla nucleata. Biol. Bull. 2003, 204, 210–214. [Google Scholar] [CrossRef]
- Bertucci, A.; Tambutte, E.; Tambutte, S.; Allemand, D.; Zoccola, D. Symbiosis-dependent gene expression in coral-dinoflagellate association: Cloning and characterization of a P-type H+-ATPase gene. Proc. Biol. Sci. 2010, 277, 87–95. [Google Scholar] [CrossRef]
- Leggat, W.; Seneca, F.; Wasmund, K.; Ukani, L.; Yellowlees, D.; Ainsworth, T.D. Differential responses of the coral host and their algal symbiont to thermal stress. PLoS One 2011, 6, e26687. [Google Scholar]
- Wohlrab, S.; Iversen, M.H.; John, U. A molecular and co-evolutionary context for grazer induced toxin production in Alexandrium tamarense. PLoS One 2010, 5, e15039. [Google Scholar]
- Yang, E.; van Nimwegen, E.; Zavolan, M.; Rajewsky, N.; Schroeder, M.; Magnasco, M.; Darnell, J.E., Jr. Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 2003, 13, 1863–1872. [Google Scholar]
- Kinniburgh, A.J.; Mertz, J.E.; Ross, J. The precursor of mouse beta-globin messenger RNA contains two intervening RNA sequences. Cell 1978, 14, 681–693. [Google Scholar] [CrossRef]
- Chow, L.T.; Gelinas, R.E.; Broker, T.R.; Roberts, R.J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 1977, 12, 1–8. [Google Scholar] [CrossRef]
- Berget, S.M.; Moore, C.; Sharp, P.A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA 1977, 74, 3171–3175. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, S. Complex gene structure of the form II Rubisco in the dinoflagellate Prorocentrum minimum (Dinophyceae). J. Phycol. 2003, 39, 1160–1171. [Google Scholar] [CrossRef]
- Rowan, R.; Whitney, S.M.; Fowler, A.; Yellowlees, D. Rubisco in marine symbiotic dinoflagellates: Form II enzymes in eukaryotic oxygenic phototrophs encoded by a nuclear multigene family. Plant Cell 1996, 8, 539–553. [Google Scholar]
- Orr, R.J.; Stuken, A.; Murray, S.A.; Jakobsen, K.S. Evolutionary acquisition and loss of saxitoxin biosynthesis in dinoflagellates: The second “core” gene, sxtG. Appl. Environ. Microbiol. 2013, 79, 2128–2136. [Google Scholar] [CrossRef]
- Kitamura-Abe, S.; Itoh, H.; Washio, T.; Tsutsumi, A.; Tomita, M. Characterization of the splice sites in GT-AG and GC-AG introns in higher eukaryotes using full-length cDNAs. J. Bioinform. Comput. Biol. 2004, 2, 309–331. [Google Scholar] [CrossRef]
- Nilsen, T.W. The spliceosome: No assembly required? Mol. Cell 2002, 9, 8–9. [Google Scholar] [CrossRef]
- Reddy, R.; Spector, D.; Henning, D.; Liu, M.H.; Busch, H. Isolation and partial characterization of dinoflagellate U1–U6 small RNAs homologous to rat U small nuclear RNAs. J. Biol. Chem. 1983, 258, 13965–13969. [Google Scholar]
- Alverca, E.; Franca, S.; Diaz de la Espina, S.M. Topology of splicing and snRNP biogenesis in dinoflagellate nuclei. Biol. Cell 2006, 98, 709–720. [Google Scholar] [CrossRef]
- Zhang, H.; Hou, Y.; Miranda, L.; Campbell, D.A.; Sturm, N.R.; Gaasterland, T.; Lin, S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 4618–4623. [Google Scholar]
- Boothroyd, J.C.; Cross, G.A. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene 1982, 20, 281–289. [Google Scholar] [CrossRef]
- Agabian, N. Trans splicing of nuclear pre-mRNAs. Cell 1990, 61, 1157–1160. [Google Scholar] [CrossRef]
- Douris, V.; Telford, M.J.; Averof, M. Evidence for multiple independent origins of trans-splicing in Metazoa. Mol. Biol. Evol. 2010, 27, 684–693. [Google Scholar] [CrossRef]
- Hastings, K.E. SL trans-splicing: Easy come or easy go? Trends Genet. 2005, 21, 240–247. [Google Scholar] [CrossRef]
- Lasda, E.L.; Blumenthal, T. Trans-splicing. Wiley Interdiscip. Rev. RNA 2011, 2, 417–434. [Google Scholar] [CrossRef]
- Vandenberghe, A.E.; Meedel, T.H.; Hastings, K.E. mRNA 5′-leader trans-splicing in the chordates. Genes Dev. 2001, 15, 294–303. [Google Scholar] [CrossRef]
- Davis, R.E. Surprising diversity and distribution of spliced leader RNAs in flatworms. Mol. Biochem. Parasitol. 1997, 87, 29–48. [Google Scholar] [CrossRef]
- Jackson, C.J.; Waller, R.F. A widespread and unusual RNA trans-splicing type in dinoflagellate mitochondria. PLoS One 2013, 8, e56777. [Google Scholar] [CrossRef]
- Hearne, J.L.; Pitula, J.S. Identification of two spliced leader RNA transcripts from Perkinsus marinus. J. Eukaryot. Microbiol. 2011, 58, 266–268. [Google Scholar]
- Slamovits, C.H.; Keeling, P.J. Widespread recycling of processed cDNAs in dinoflagellates. Curr. Biol. 2008, 18, R550–R552. [Google Scholar] [CrossRef]
- Zhang, H.; Dungan, C.F.; Lin, S. Introns, alternative splicing, spliced leader trans-splicing and differential expression of pcna and cyclin in Perkinsus marinu. Protist 2011, 162, 154–167. [Google Scholar] [CrossRef]
- Suntharalingam, M.; Wente, S.R. Peering through the pore: Nuclear pore complex structure, assembly, and function. Dev. Cell 2003, 4, 775–789. [Google Scholar] [CrossRef]
- Vasu, S.K.; Forbes, D.J. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 2001, 13, 363–375. [Google Scholar] [CrossRef]
- Fried, H.; Kutay, U. Nucleocytoplasmic transport: Taking an inventory. Cell. Mol. Life Sci. 2003, 60, 1659–1688. [Google Scholar] [CrossRef]
- Frankel, M.B.; Knoll, L.J. The ins and outs of nuclear trafficking: Unusual aspects in apicomplexan parasites. DNA Cell Biol. 2009, 28, 277–284. [Google Scholar] [CrossRef]
- Kohler, A.; Hurt, E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007, 8, 761–773. [Google Scholar]
- Zhang, J.; Sun, X.; Qian, Y.; LaDuca, J.P.; Maquat, L.E. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: A possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 1998, 18, 5272–5283. [Google Scholar]
- Zhang, J.; Sun, X.; Qian, Y.; Maquat, L.E. Intron function in the nonsense-mediated decay of beta-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 1998, 4, 801–815. [Google Scholar] [CrossRef]
- Isken, O.; Maquat, L.E. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev. 2007, 21, 1833–1856. [Google Scholar] [CrossRef]
- Ito-Harashima, S.; Kuroha, K.; Tatematsu, T.; Inada, T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007, 21, 519–524. [Google Scholar] [CrossRef]
- Wu, S.; Wang, W.; Kong, X.; Congdon, L.M.; Yokomori, K.; Kirschner, M.W.; Rice, J.C. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010, 24, 2531–2542. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Roy, S.; Morse, D. Transcription and Maturation of mRNA in Dinoflagellates. Microorganisms 2013, 1, 71-99. https://doi.org/10.3390/microorganisms1010071
Roy S, Morse D. Transcription and Maturation of mRNA in Dinoflagellates. Microorganisms. 2013; 1(1):71-99. https://doi.org/10.3390/microorganisms1010071
Chicago/Turabian StyleRoy, Sougata, and David Morse. 2013. "Transcription and Maturation of mRNA in Dinoflagellates" Microorganisms 1, no. 1: 71-99. https://doi.org/10.3390/microorganisms1010071
APA StyleRoy, S., & Morse, D. (2013). Transcription and Maturation of mRNA in Dinoflagellates. Microorganisms, 1(1), 71-99. https://doi.org/10.3390/microorganisms1010071





