Opportunistic Pathogens Mycobacterium Avium Complex (MAC) and Legionella spp. Colonise Model Shower
Abstract
:1. Introduction
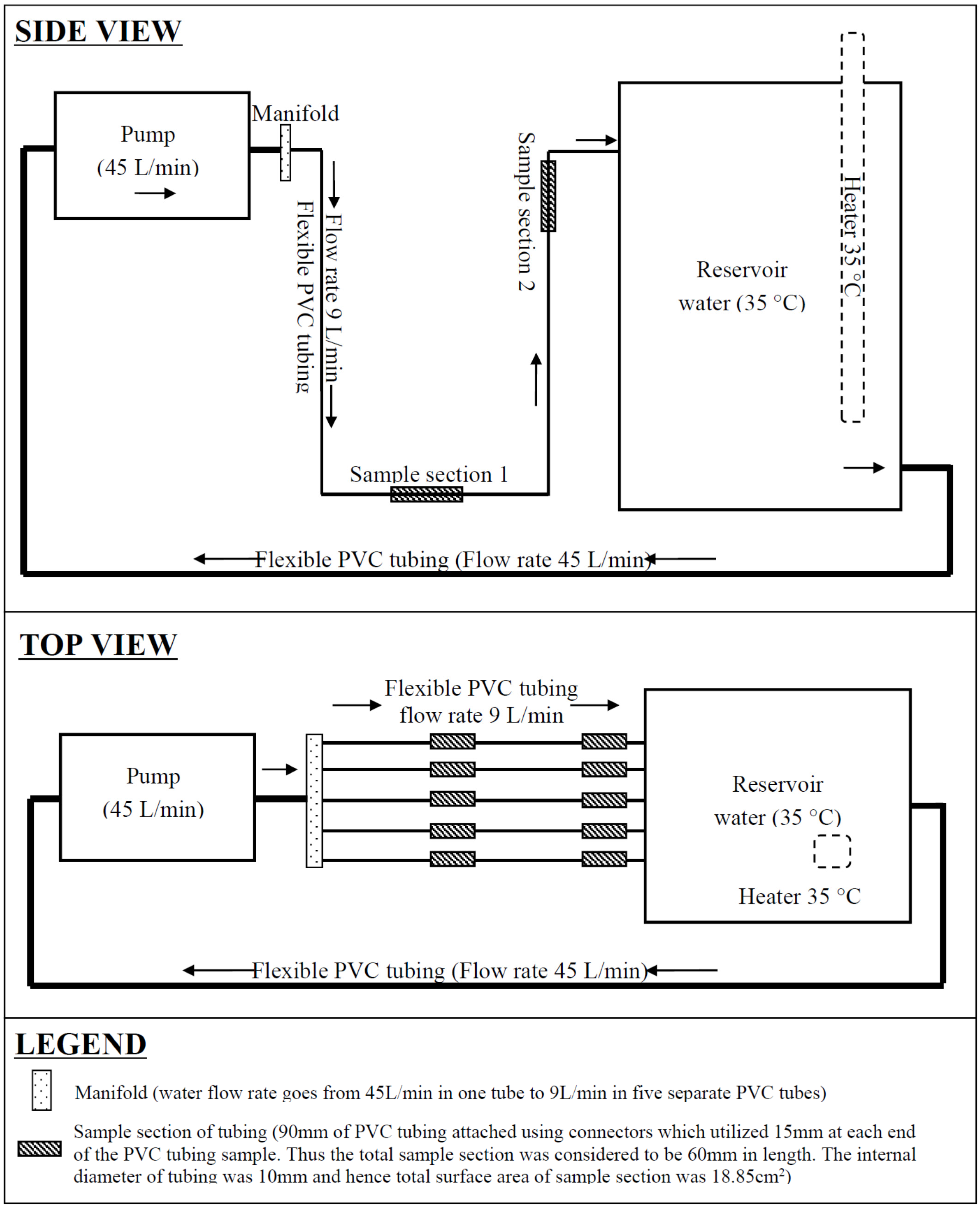
2. Experimental Model and Methods
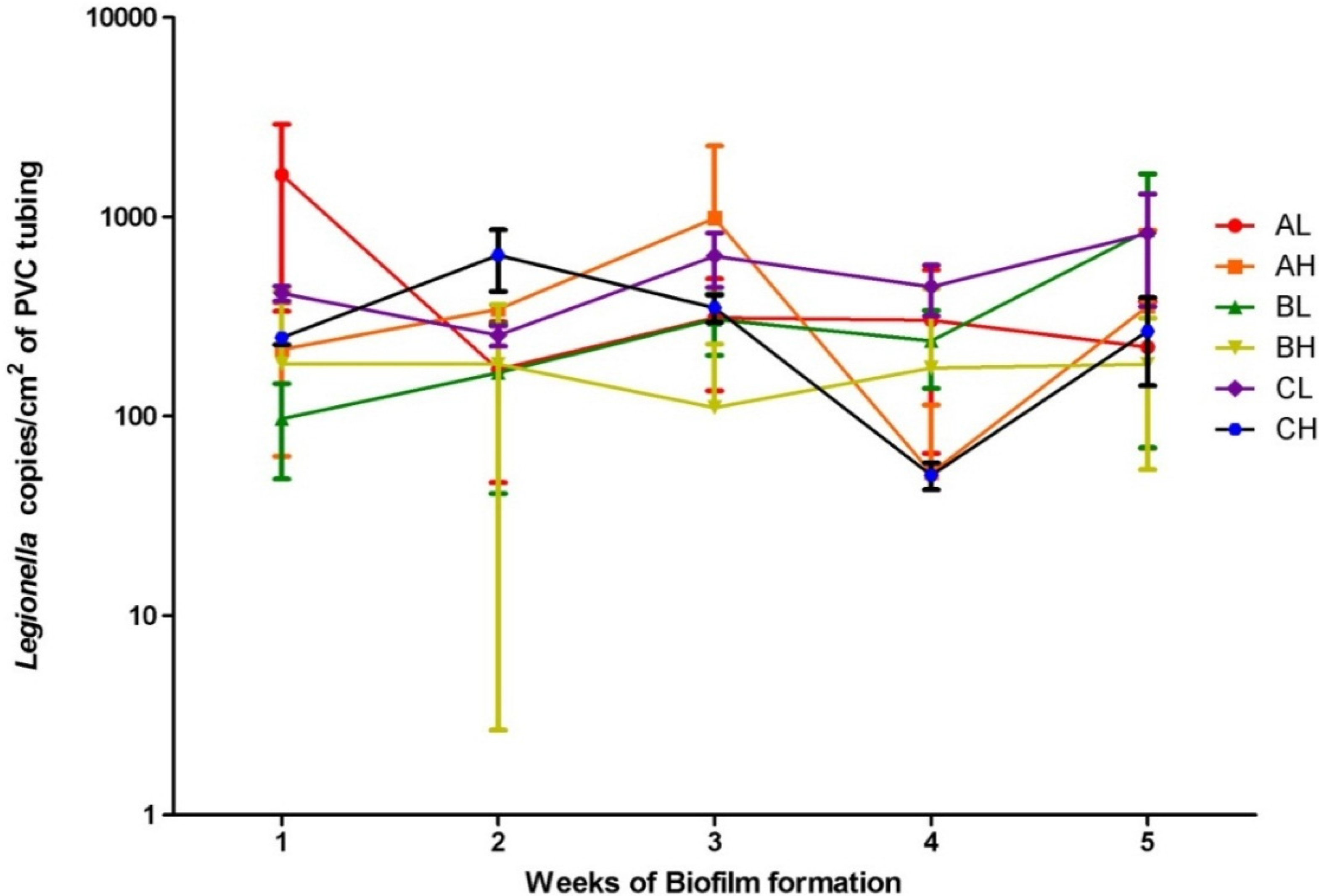
3. Results and Discussion

4. Conclusions
Author Contributions
Conflicts of Interest
References
- Lindsay, D.; Holy, A. Bacterial biofilms within the clinical setting: What healthcare professionals should know. J. Hosp. infect. 2006, 64, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Feazel, L.M.; Baumgartner, L.K.; Peterson, K.L.; Frank, D.N.; Harris, J.K.; Pace, N.R. Opportunistic pathogens enriched in showerhead biofilms. Proc. Natl. Acad. Sci. 2009, 106, 16393–16399. [Google Scholar] [CrossRef] [PubMed]
- Zmirou-Navier, D.; Remen, T.; Bauer, M.; Deloge-Abarkan, M.; Tossa, P.; Mathieu, L. Legionella in shower aerosols and pontiac fever among health workers and residents in nursing homes. Epidemiology 2007, 18, S50. [Google Scholar] [CrossRef]
- Falkinham, J.O.; Iseman, M.D.; de Haas, P.; van Soolingen, D. Mycobacterium avium in a shower linked to pulmonary disease. J. Water Health 2008, 6, 209–213. [Google Scholar] [PubMed]
- Field, S.K. Mycobacterium avium complex pulmonary disease in patients without hiv infection. Chest 2004, 126, 566. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.H.; Kao, P.N.; Adi, V.; Ruoss, S.J. Mycobacterium avium-intracellulare pulmonary infection in hiv-negative patients without preexisting lung disease. Chest 1999, 115, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Lakhanpal, A.; Arfon, S.; McKeon, D.J. So, they thought it was all over. BMJ Case Rep. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Marras, T.K.; Wallace, R.J.; Koth, L.L.; Stulbarg, M.S.; Cowl, C.T.; Daley, C.L. Hypersensitivity pneumonitis reaction to mycobacterium avium in household water. Chest 2005, 127, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Sugita, Y. Familial cluster of cutaneous mycobacterium avium infection resulting from use of a circulating, constantly heated bath water system. Br. J. Dermatol. 2000, 142, 789. [Google Scholar] [CrossRef] [PubMed]
- Karakousis, P.C.; Moore, R.D.; Chaisson, R.E. Mycobacterium avium complex in patients with hiv infection in the era of highly active antiretroviral therapy. The Lancet 2004, 4, 557–565. [Google Scholar] [CrossRef]
- Thegerström, J.; Friman, V.; Nylén, O.; Romanus, V.; Olsen, B. Clinical features and incidence of mycobacterium avium infections in children. Scand. J. Infect. Dis. 2008, 40, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Nightingale, S.D.; Byrd, L.T.; Southern, P.M.; Jockusch, J.D.; Cal, S.X.; Wynne, B.A. Incidence of mycobacterium avium-intracellulare complex bacteremia in human immunodeficiency virus (hiv) positive patients. J. Infect. Dis. 1992, 165, 1082–1085. [Google Scholar] [CrossRef] [PubMed]
- Naser, S.A.; Ghobrial, G.; Romero, C.; Valentine, J.F. Culture of mycobacterium avium subspecies paratuberculosis from the blood of patients with crohn’s disease. The Lancet 2004, 364, 1039–1044. [Google Scholar] [CrossRef]
- Adjemian, J.; Olivier, K.N.; Seitz, A.E.; Holland, S.M.; Prevots, D.R. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am. J. Respir. Crit. Care Med. 2012, 185, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Al-Houqani, M.; Jamieson, F.; Mehta, M.; Chedore, P.; May, K.; Marras, T.K. Aging, copd, and other risk factors do not explain the increased prevalence of pulmonary mycobacterium avium complex in ontario. Chest 2012, 141, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Marras, T.K.; Daley, C.L. Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin. Chest Med. 2002, 23, 553–567. [Google Scholar] [CrossRef]
- O’Brien, D.P.; Currie, B.J.; Krause, V.L. Nontuberculous mycobacterial disease in northern australia: A case series and review of the literature. Clin. Infect. Dis. 2000, 31, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Nishiuchi, Y.; Maekura, R.; Kitada, S.; Tamaru, A.; Taguri, T.; Kira, Y.; Hiraga, T.; Hirotani, A.; Yoshimura, K.; Miki, M.; et al. The recovery of mycobacterium avium-intracellulare complex (mac) from the residential bathrooms of patients with pulmonary mac. Clin. Infect. Dis. 2007, 45, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, S.; Trebesius, K.; Heesemann, Jr. Evalution of detection of legionella spp. In water samples by fluorescence in situ hybridization, pcr amplification and bacterial culture. Int. J. Medical Microbiol. 2002, 292, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Pond, K. Water recreation and disease. In Plausibility of Associated Infections: Aute Effects, Sequelae and Mortality; IWA Publishing on behalf of World Health Organization (WHO): London, UK, 2005. [Google Scholar]
- Kool, J.; Bergmire-Sweat, D.; Butler, J.; Brown, E.; Peabody, D.; Massi, D.; Carpenter, J.; Pruckler, J.; Robert, B.; Fields, B. Hospital characteristics associated with colonization of water systems by legionella and risk of nosocomial legionnaires’ disease: A cohort study of 15 hospitals. Infect. Control Hosp. Epidemiol. 1999, 20, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Wellinghausen, N.; Frost, C.; Marre, R. Detection of legionellae in hospital water samples by quantitative real-time lightcycler pcr. Appl. Environ. Microbiol. 2001, 67, 3985–3993. [Google Scholar] [CrossRef] [PubMed]
- Cordes, L.; Wiesenthal, A.; Gorman, G.; Phair, J.; Sommers, H.; Brown, A.; Yu, V.; Magnussen, M.; Meyer, R.; Wolf, J.; et al. Isolation of legionella pneumophila from hospital shower heads. Ann. Intern. Med. 1981, 94, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Whiley, H.; Taylor, M. Legionella detection by culture and qpcr: Comparing apples and oranges. Crit. Rev. Microbiol. 2014. [Google Scholar]
- Eaton, T.; Falkinham, J.O.; Aisu, T.O.; Daniel, T.M. Isolation and characteristics of mycobacterium avium complex from water and soil samples in uganda. Tuber. Lung Dis. 1995, 76, 570–574. [Google Scholar] [CrossRef]
- Han, X.Y.; Tarrand, J.J.; Infante, R.; Jacobson, K.L.; Truong, M. Clinical significance and epidemiologic analyses of mycobacterium avium and mycobacterium intracellulare among patients without aids. J. Clin. Microbiol. 2005, 43, 4407–4412. [Google Scholar] [CrossRef] [PubMed]
- Giglio, S.; Monis, P.T.; Saint, C.P. Legionella confirmation using real-time pcr and syto9 is an alternative to current methodology. Appl. Environ. Microbiol. 2005, 71, 8944–8948. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Jang, H.; Kim, C.; Chung, B.; Chang, C.; Park, S.K.; Song, S. Detection and identification of mycobacteria by amplification of the internal transcribed spacer regions with genus- and species-specific pcr primers. J. Clin. Microbiol. 2000, 38, 4080. [Google Scholar] [PubMed]
- Yáñez, M.A.; Nocker, A.; Soria-Soria, E.; Múrtula, R.; Martínez, L.; Catalán, V. Quantification of viable legionella pneumophila cells using propidium monoazide combined with quantitative pcr. J. Microbiol. Methods 2011, 85, 124–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Whiley, H.; Giglio, S.; Bentham, R. Opportunistic Pathogens Mycobacterium Avium Complex (MAC) and Legionella spp. Colonise Model Shower. Pathogens 2015, 4, 590-598. https://doi.org/10.3390/pathogens4030590
Whiley H, Giglio S, Bentham R. Opportunistic Pathogens Mycobacterium Avium Complex (MAC) and Legionella spp. Colonise Model Shower. Pathogens. 2015; 4(3):590-598. https://doi.org/10.3390/pathogens4030590
Chicago/Turabian StyleWhiley, Harriet, Steven Giglio, and Richard Bentham. 2015. "Opportunistic Pathogens Mycobacterium Avium Complex (MAC) and Legionella spp. Colonise Model Shower" Pathogens 4, no. 3: 590-598. https://doi.org/10.3390/pathogens4030590





