Epidemiological Investigation of Tick-Borne Bacterial Pathogens in Domestic Animals from the Qinghai–Tibetan Plateau Area, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Blood Sample Collection
2.2. Morphological and Molecular Identification of Ticks
2.3. DNA Extraction
2.4. Molecular Identification of Tick-Borne Bacterial Pathogens
2.5. Sequencing Analyses of Detected Tick-Borne Bacterial Pathogens
2.6. Phylogenetic Tree of Detected Tick-Borne Bacterial Pathogens
2.7. Statistical Analyses
3. Results
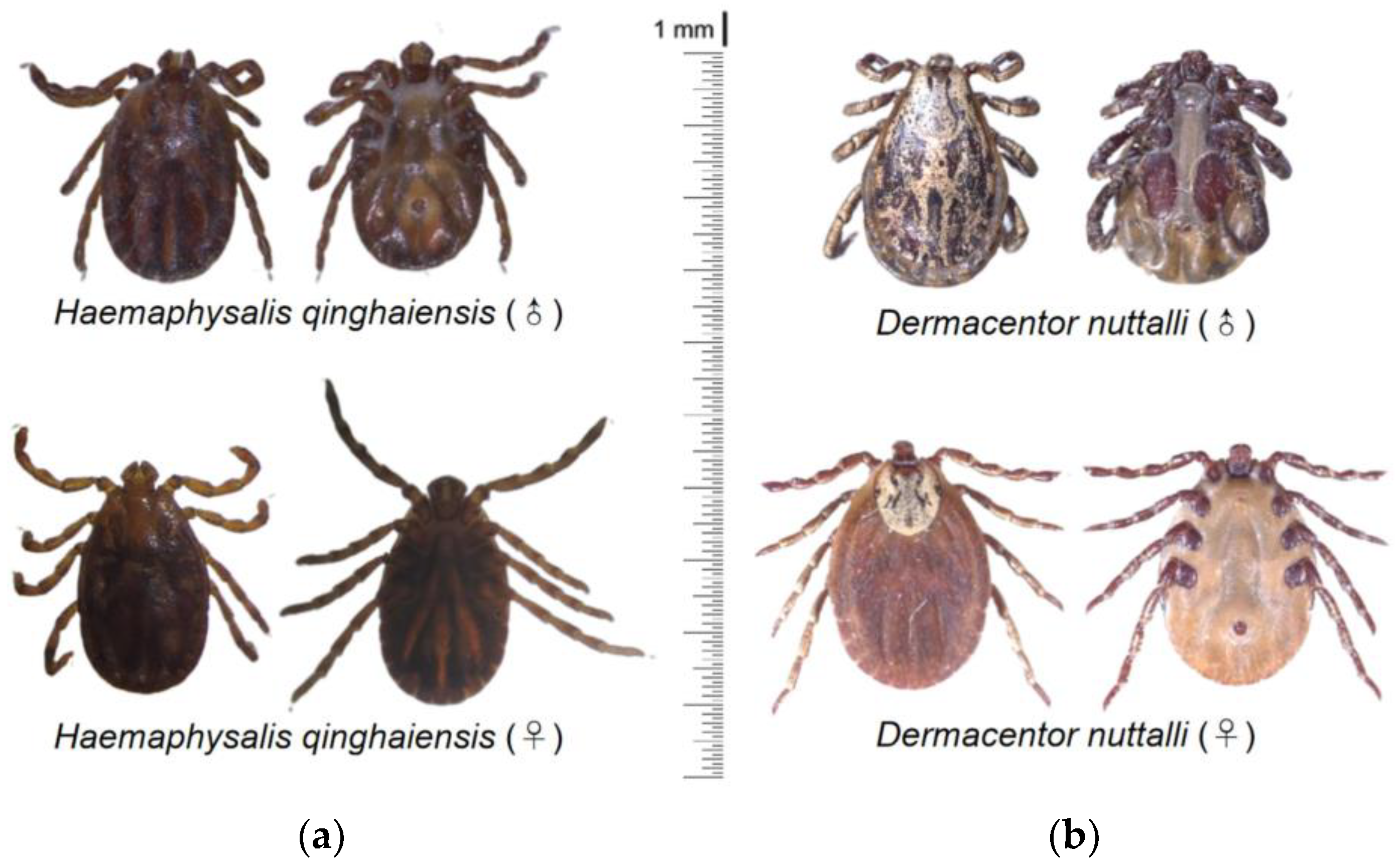
3.1. Morphological and Molecular Identification of Ticks
3.2. Identification of Tick-Borne Bacterial Pathogens
3.3. Sequencing and Comparative Analyses
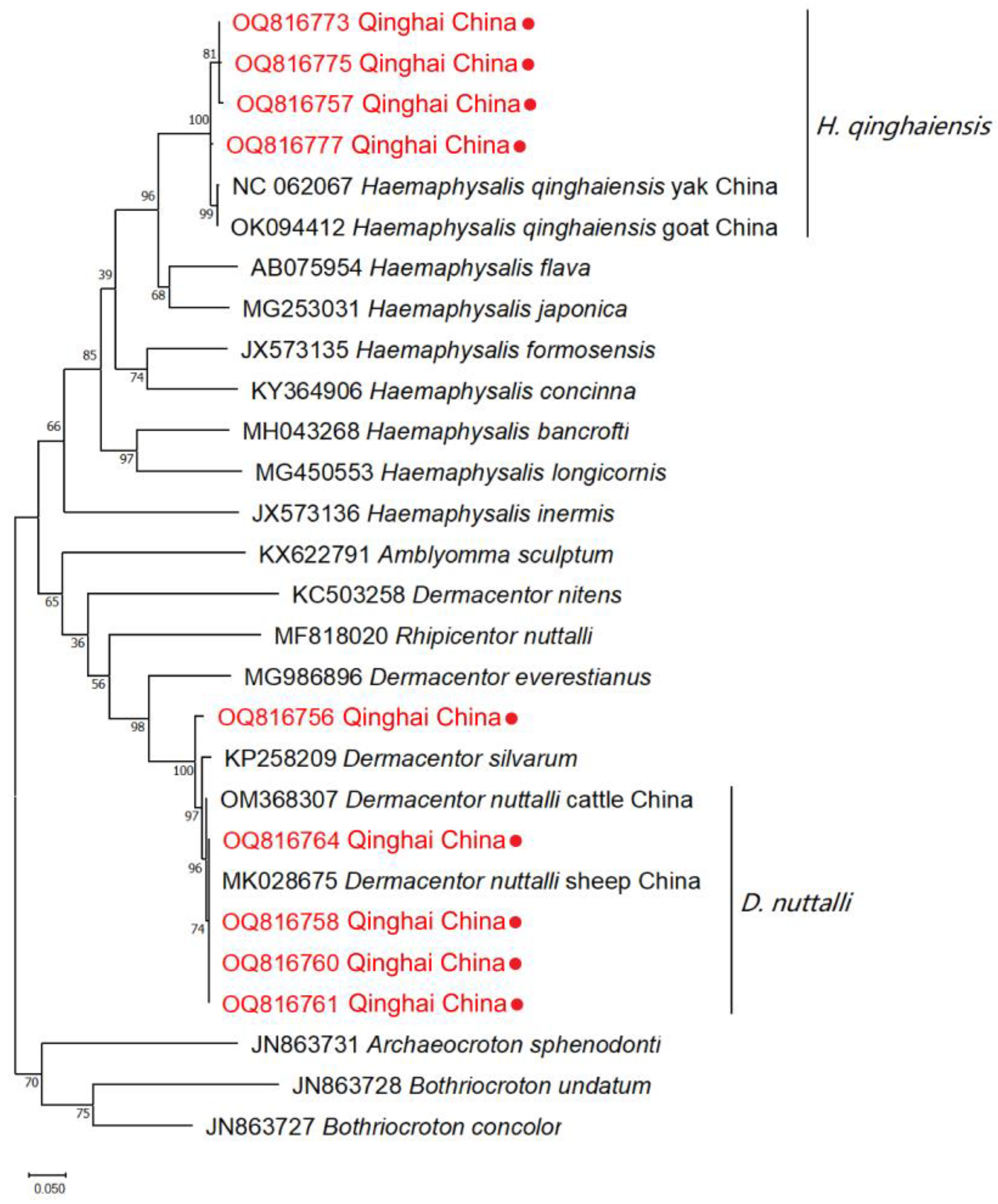
3.4. Phylogenetic Analyses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, Z.; Li, L.; Hu, Y.; Hu, H.; Li, C.; Ping, X.; Luo, Z. Diversity and endemism of ungulates on the Qinghai-Tibetan plateau: Evolution and conservation. Biodivers. Sci. 2018, 26, 158–170. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Li, Q.; Tseng, Z.J.; Takeuchi, G.T.; Deng, T.; Xie, G.; Chang, M.; Wang, N. Cenozoic vertebrate evolution and paleoenvironment in Tibetan plateau: Progress and prospects. Gondwana Res. 2015, 27, 1335–1354. [Google Scholar] [CrossRef]
- Jin, Z.; Zhuang, Q.; He, J.-S.; Luo, T.; Shi, Y. Phenology shift from 1989 to 2008 on the Tibetan plateau: An analysis with a process-based soil physical model and remote sensing data. Clim. Chang. 2013, 119, 435–449. [Google Scholar] [CrossRef]
- Spicer, R.A.; Su, T.; Valdes, P.J.; Farnsworth, A.; Wu, F.-X.; Shi, G.; Spicer, T.E.V.; Zhou, Z. Why ‘the Uplift of the Tibetan Plateau’ Is a Myth? Natl. Sci. Rev. 2020, 8, nwaa091. [Google Scholar]
- Qi, T.; Ai, J.; Yang, J.; Zhu, H.; Zhou, Y.; Zhu, Y.; Zhang, H.; Qin, Q.; Kang, M.; Sun, Y.; et al. Seroepidemiology of neosporosis in various animals in the Qinghai-Tibetan plateau. Front. Vet. Sci. 2022, 9, 953380. [Google Scholar] [CrossRef]
- Anderson, J.F.; Magnarelli, L.A. Biology of ticks. Infect. Dis. Clin. N. Am. 2008, 22, 195–215. [Google Scholar] [CrossRef]
- Moraga-Fernández, A.; Muñoz-Hernández, C.; Sánchez-Sánchez, M.; de Mera IG, F.; de la Fuente, J. Exploring the diversity of tick-borne pathogens: The case of bacteria (Anaplasma, Rickettsia, Coxiella and Borrelia) protozoa (Babesia and Theileria) and viruses (Orthonairovirus, tick-borne encephalitis virus and louping ill virus) in the European continent. Vet. Microbiol. 2023, 286, 109892. [Google Scholar] [CrossRef]
- Wang, Y.; Han, S.; He, H. Research status of tick distribution and tick-borne diseases in Qinghai province. Prev. Vet. Med. 2022, 43, 115–119. [Google Scholar]
- Li, H.; Zheng, Y.C.; Ma, L.; Jia, N.; Jiang, B.G.; Jiang, R.R.; Huo, Q.B.; Wang, Y.W.; Liu, H.B.; Chu, Y.L.; et al. Human infection with a novel tick-borne Anaplasma species in China: A surveillance study. Infect. Dis. 2015, 15, 663–670. [Google Scholar] [CrossRef]
- Zhou, Z.; Nie, K.; Tang, C.; Wang, Z.; Zhou, R.; Hu, S.; Zhang, Z. Phylogenetic analysis of the genus Anaplasma in southwestern China based on 16S rRNA sequence. Res. Vet. Sci. 2010, 89, 262–265. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, M.; Wang, Z.; Wang, J.; Peng, Y.; Li, Y.; Guan, G.; Luo, J.; Yin, H. Molecular survey and genetic identification of Anaplasma species in goats from central and southern China. Appl. Environ. Microbiol. 2012, 78, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, Y.; Liu, Z.; Liu, J.; Niu, Q.; Ren, Q.; Chen, Z.; Guan, G.; Luo, J.; Yin, H. Molecular detection and characterization of Anaplasma spp. in sheep and cattle from Xinjiang, northwest China. Parasites Vectors 2015, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Z.; Niu, Q.; Liu, J.; Guan, G.; Xie, J.; Luo, J.; Wang, S.; Wang, S.; Yin, H. First molecular survey and identification of Anaplasma spp. in white yaks (Bos grunniens) in China. Parasitology 2016, 143, 686–691. [Google Scholar] [PubMed]
- Jian, Y.; Zhang, X.; Li, X.; Wang, G.; Wang, G.; Ma, L. Identification of ticks and tick-borne pathogens in different areas of Haibei of Qinghai province. Chin. Qinghai J. Anim. Vet. Sci. 2020, 50, 35. [Google Scholar]
- Wang, G.; van Dam, A.P.; Schwartz, I.; Dankert, J. Molecular Typing of Borrelia burgdorferi sensu lato: Taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 1999, 12, 633–653. [Google Scholar] [CrossRef]
- Hao, Q.; Hou, X.; Geng, Z.; Wan, K. Distribution of Borrelia burgdorferi sensu lato in China. J. Clin. Microbiol. 2011, 49, 647–650. [Google Scholar] [PubMed]
- Yue, J.; Shi, Y. Epidemiological investigation of Lyme disease in parts of forest areas in Qinghai province. Chin. J. Vector. Biol. Control 2009, 20, 358–359. [Google Scholar]
- Han, R. Studies on Species Diversity of Ticks and Gene Polymorphism of Tick-Borne Pathogens in Qinghai Province. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Lanzhou, China, 2018. [Google Scholar]
- Jian, Y.; Li, J.; Adjou Moumouni, P.F.; Zhang, X.; Tumwebaze, M.A.; Wang, G.; Cai, Q.; Li, X.; Wang, G.; Liu, M.; et al. Human spotted fever group Rickettsia infecting yaks (Bos grunniens) in the Qinghai-Tibetan plateau area. Pathogens 2020, 9, 249. [Google Scholar]
- Yu, X.; Jin, Y.; Fan, M.; Xu, G.; Liu, Q.; Raoult, D. Genotypic and antigenic identification of two new strains of spotted fever group rickettsiae isolated from China. J. Clin. Microbiol. 1993, 31, 83–88. [Google Scholar] [CrossRef]
- Li, W.; Liu, L.; Jiang, X.; Guo, X.; Garnier, M.; Raoult, D.; Parola, P. Molecular identification of spotted fever group rickettsiae in ticks collected in central China. Clin. Microbiol. Infect. 2009, 15, 279–280. [Google Scholar] [CrossRef]
- Tian, Z.C.; Liu, G.Y.; Shen, H.; Xie, J.R.; Luo, J.; Tian, M.Y. First Report on the occurrence of Rickettsia slovaca and Rickettsia raoultii in Dermacentor silvarum in China. Parasites Vectors 2012, 5, 19. [Google Scholar] [PubMed]
- Zhang, J.; Lu, G.; Kelly, P.; Zhang, Z.; Wei, L.; Yu, D.; Kayizha, S.; Wang, C. First report of Rickettsia felisin China. BMC Infect. Dis. 2014, 14, 682. [Google Scholar]
- Wei, Q.Q.; Guo, L.P.; Wang, A.D.; Mu, L.M.; Zhang, K.; Chen, C.F.; Zhang, W.J.; Wang, Y.Z. The first detection of Rickettsia aeschlimannii and Rickettsia massiliae in Rhipicephalus turanicus ticks, in northwest China. Parasites Vectors 2015, 8, 631. [Google Scholar] [CrossRef]
- Ge, Y.; Yin, H.; Rikihisa, Y.; Pan, W.; Yin, H. Molecular detection of tick-borne Rickettsiales in goats and sheep from southeastern China. Vector Borne Zoonotic Dis. 2016, 16, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, Q.; Zhang, X.; Li, Z.; Wang, Z.; Song, M.; Wei, F.; Wang, S.; Liu, Q. Characterization of Rickettsiae in ticks in northeastern China. Parasites Vectors 2016, 9, 498. [Google Scholar] [CrossRef]
- Han, R.; Yang, J.; Niu, Q.; Liu, Z.; Chen, Z.; Kan, W.; Hu, G.; Liu, G.; Luo, J.; Yin, H. Molecular prevalence of spotted fever group Rickettsiae in ticks from Qinghai province, northwestern China. Infect. Genet. Evol. 2018, 57, 1–7. [Google Scholar] [CrossRef]
- Gao, Y.; Li, S.; Wang, L.; Wei, F. Epidemiological investigation of tick-transmitted Rickettsia in Qinghai province. Heilongjiang Anim. Sci. Vet. Med. 2019, 18, 65–69. [Google Scholar]
- Li, J.; Jian, Y.; Jia, L.; Galon, E.M.; Benedicto, B.; Wang, G.; Cai, Q.; Liu, M.; Li, Y.; Ji, S.; et al. Molecular characterization of tick-borne bacteria and protozoans in yaks (Bos grunniens), Tibetan sheep (Ovis aries) and Bactrian camels (Camelus bactrianus) in the Qinghai-Tibetan plateau area, China. Ticks Tick Borne Dis. 2020, 11, 101466. [Google Scholar] [CrossRef]
- Maurin, M.; Raoult, D. Q fever. Clin. Microbiol. Rev. 1999, 12, 518–553. [Google Scholar]
- El-mahallawy, H.S.; Lu, G.; Kelly, P.; Xu, D.; Li, Y.; Fan, W.; Wang, C. Q fever in China: A systematic review, 1989–2013. Epidemiol. Infect. 2014, 143, 673–681. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Liu, Z.; Ren, Q.; Ma, M.; Luo, J.; Yin, H. Scanning electron microscopy of all parasitic stages of Haemaphysalis qinghaiensis Teng, 1980 (Acari: Ixodidae). Parasitol. Res. 2014, 113, 2095–2102. [Google Scholar] [CrossRef] [PubMed]
- Chitimia, L.; Lin, R.-Q.; Cosoroaba, I.; Wu, X.-Y.; Song, H.-Q.; Yuan, Z.-G.; Zhu, X.-Q. Genetic characterization of ticks from southwestern Romania by sequences of mitochondrial cox1 and nad5 genes. Exp. Appl. Acarol. 2010, 52, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, L.; Moumouni, P.F.A.; Jian, Y.; Wang, G.; Zhang, X.; Li, X.; Wang, G.; Lee, S.-H.; Galon, E.M.; et al. Molecular survey and characterization of tick-borne pathogens in sheep from Qinghai, China. Small Rumin. Res. 2019, 175, 23–30. [Google Scholar] [CrossRef]
- Barlough, J.E.; Madigan, J.E.; DeRock, E.; Bigornia, L. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus). Vet. Parasitol. 1996, 63, 319–329. [Google Scholar] [CrossRef]
- Kawahara, M.; Rikihisa, Y.; Lin, Q.; Isogai, E.; Tahara, K.; Itagaki, A.; Hiramitsu, Y.; Tajima, T. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl. Environ. Microbiol. 2006, 72, 1102–1109. [Google Scholar]
- Torina, A.; Agnone, A.; Blanda, V.; Alongi, A.; D’Agostino, R.; Caracappa, S.; Marino, A.M.F.; Di Marco, V.; de la Fuente, J. Development and validation of two PCR tests for the detection of and differentiation between Anaplasma ovis and Anaplasma marginale. Ticks Tick Borne Dis. 2012, 3, 283–287. [Google Scholar]
- Massung, R.F.; Slater, K.; Owens, J.H.; Nicholson, W.L.; Mather, T.N.; Solberg, V.B.; Olson, J.G. Nested PCR assay for detection of granulocytic Ehrlichiae. J. Clin. Microbiol. 1998, 36, 1090–1095. [Google Scholar] [CrossRef]
- Ni, X.-B.; Jia, N.; Jiang, B.-G.; Sun, T.; Zheng, Y.-C.; Huo, Q.-B.; Liu, K.; Ma, L.; Zhao, Q.-M.; Yang, H.; et al. Lyme borreliosis caused by diverse genospecies of Borrelia burgdorferi sensu lato in northeastern China. Clin. Microbiol. Infect. 2014, 20, 808–814. [Google Scholar] [CrossRef]
- Zhai, B.; Niu, Q.; Yang, J.; Liu, Z.; Liu, J.; Yin, H.; Zeng, Q. Identification and molecular survey of Borrelia burgdorferi sensu lato in sika deer (Cervus nippon) from Jilin province, north-eastern China. Acta Trop. 2017, 166, 54–57. [Google Scholar] [CrossRef]
- To, H.; Kako, N.; Zhang, G.Q.; Otsuka, H.; Ogawa, M.; Ochiai, O.; Nguyen, S.V.; Yamaguchi, T.; Fukushi, H.; Nagaoka, N.; et al. Q fever pneumonia in Children in Japan. J. Clin. Microbiol. 1996, 34, 647–651. [Google Scholar] [CrossRef]
- Kidd, L.; Maggi, R.; Diniz, P.P.V.P.; Hegarty, B.; Tucker, M.; Breitschwerdt, E. Evaluation of conventional and real-time PCR assays for detection and differentiation of spotted fever group Rickettsia in dog blood. Vet. Microbiol. 2008, 129, 294–303. [Google Scholar] [CrossRef]
- Roux, V.; Raoult, D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (OmpB). Int. J. Syst. Evol. Microbiol. 2000, 50, 1449–1455. [Google Scholar] [CrossRef] [PubMed]
- Labruna, M.B.; Whitworth, T.; Horta, M.C.; Bouyer, D.H.; McBride, J.W.; Pinter, A.; Popov, V.; Gennari, S.M.; Walker, D.H. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of São Paulo, Brazil, where Brazilian spotted fever is endemic. J. Clin. Microbiol. 2004, 42, 90–98. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [PubMed]
- Han, R.; Yang, J.-F.; Mukhtar, M.U.; Chen, Z.; Niu, Q.-L.; Lin, Y.-Q.; Liu, G.-Y.; Luo, J.-X.; Yin, H.; Liu, Z.-J. Molecular detection of Anaplasma Infections in Ixodid ticks from the Qinghai-Tibet plateau. Infect. Dis. Poverty 2019, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Chen, W.; Ma, P.; Wei, Y.; Li, R.; Chen, Z.; Tian, S.; Qi, T.; Yang, J.; Sun, Y.; et al. Molecular detection of Anaplasma spp., Babesia spp. and Theileria spp. in yaks (Bos grunniens) and Tibetan sheep (Ovis aries) on the Qinghai-Tibetan Plateau, China. Parasites Vectors 2021, 14, 613. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, J.; Atkinson, M.W.; Naranjo, V.; Fernández de Mera, I.G.; Mangold, A.J.; Keating, K.A.; Kocan, K.M. Sequence analysis of the msp4 gene of Anaplasma ovis strains. Vet. Microbiol. 2007, 119, 375–381. [Google Scholar] [CrossRef]
- Del Cerro, A.; Oleaga, A.; Somoano, A.; Barandika, J.F.; García-Pérez, A.L.; Espí, A. Molecular identification of tick-borne pathogens (Rickettsia spp., Anaplasma phagocytophilum, Borrelia burgdorferi sensu lato, Coxiella burnetii and piroplasms) in questing and feeding hard ticks from North-Western Spain. Ticks Tick-Borne Dis. 2022, 13, 101961. [Google Scholar] [CrossRef]
- Zhao, G.P.; Wang, Y.X.; Fan, Z.W.; Ji, Y.; Liu, M.; Zhang, W.H.; Li, X.L.; Zhou, S.X.; Li, H.; Liang, S.; et al. Mapping ticks and tick-borne pathogens in China. Nat. Commun. 2021, 12, 1075. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Q.; Liu, J.Q.; Xu, B.L.; Lv, S.; Xia, S.; Zhou, X.N. Tick-borne pathogens and associated co-infections in ticks collected from domestic animals in central China. Parasites Vectors 2014, 7, 237. [Google Scholar] [CrossRef]
- Wu, X.B.; Na, R.H.; Wei, S.S.; Zhu, J.S.; Peng, H.J. Distribution of tick-borne diseases in China. Parasites Vectors 2013, 6, 119. [Google Scholar] [CrossRef]
- Hirunkanokpun, S.; Ahantarig, A.; Baimai, V.; Pramual, P.; Rakthong, P.; Trinachartvanit, W. Spotted fever group Rickettsia, Anaplasma and Coxiella-like endosymbiont in Haemaphysalis ticks from mammals in Thailand. Vet. Res. Commun. 2022, 46, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Chen, Z.; Robbins, R.G. Haemaphysalis qinghaiensis (Acari: Ixodidae), a correct original species name, with notes on Chinese geographical and personal names in zoological taxa. Syst. Appl. Acarol. 2016, 21, 267. [Google Scholar]
- Yang, J.; Liu, Z.; Guan, G.; Liu, Q.; Li, Y.; Chen, Z.; Ma, M.; Liu, A.; Ren, Q.; Luo, J.; et al. Prevalence of Anaplasma phagocytophilum in ruminants, rodents and ticks in Gansu, north-western China. J. Med. Microbiol. 2013, 62, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Han, R.; Liu, Z.; Niu, Q.; Guan, G.; Liu, G.; Luo, J.; Yin, H. Insight into the Genetic Diversity of Anaplasma marginale in cattle from ten provinces of China. Parasites Vectors 2017, 10, 565. [Google Scholar] [PubMed]
- Wen, B.; Jian, R.; Zhang, Y.; Chen, R. Simultaneous detection of Anaplasma marginale and a new Ehrlichia species closely related to Ehrlichia chaffeensis by sequence analyses of 16S ribosomal DNA in Boophilus microplus ticks from Tibet. J. Clin. Microbiol. 2002, 40, 3286–3290. [Google Scholar] [CrossRef]
- Gao, Y. Distribution of tick species and epidemiological investigation of 3 species of bacterial and protozoal in Qinghai province. Master’s Thesis, Jilin Agricultural University, Changchun, China, 2018. [Google Scholar]
- Ni, J.; Lin, H.; Xu, X.; Ren, Q.; Aizezi, M.; Luo, J.; Luo, Y.; Ma, Z.; Chen, Z.; Tan, Y.; et al. Coxiella burnetii is widespread in ticks (Ixodidae) in the Xinjiang areas of China. BMC Vet. Res. 2020, 16, 317. [Google Scholar] [CrossRef]
- Yin, M.Y.; Qin, S.Y.; Tan, Q.D.; Feng, S.Y.; Liu, G.X.; Zhou, D.H.; Zhu, X.Q. First Report of Coxiella burnetii seroprevalence in Tibetan sheep in China. Vector Borne Zoonotic Dis. 2015, 15, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Shurong, Y. Coxiella burnetii in China. Q. Fever 1991, 2, 327–341. [Google Scholar]





| Species | Target Gene | Method | Primer Sequence | Annealing Temperature (°C) | Amplicon Size (bp) | Reference | |
|---|---|---|---|---|---|---|---|
| A. bovis | 16S rRNA | PCR | F | TCCTGGCTCAGAACGAACGCTGGCGGC | 55 | 1433 | [35] |
| R | AGTCACTGACCCAACCTTAAATGGCTG | ||||||
| nPCR 1 | nF 2 | CTCGTAGCTTGCTATGAGAAC | 55 | 551 | [36] | ||
| nR 3 | TCTCCCGGACTCCAGTCTG | ||||||
| A. capra | gltA | PCR | F | GCGATTTTAGAGTGYGGAGATTG | 55 | 1031 | [9] |
| R | TACAATACCGGAGTAAAAGTCAA | ||||||
| nPCR | nF | GGGTTCMTGTCYACTGCTGCGTG | 55 | 793 | |||
| nR | TTGGATCGTARTTCTTGTAGACC | ||||||
| A. marginale | msp4 | PCR | F | CTGAAGGGGGAGTAATGGG | 60 | 344 | [37] |
| R | GGTAATAGCTGCCAGAGATTCC | ||||||
| A. ovis | msp4 | PCR | F | TGAAGGGAGCGGGGTCATGGG | 62 | 347 | [37] |
| R | GAGTAATTGCAGCCAGGCACTCT | ||||||
| A. phagocytophilum | 16S rRNA | PCR | F | CACATGCAAGTCGAACGGATTATTC | 55 | 932 | [38] |
| R | TTCCGTTAAGAAGGATCTAATCTCC | ||||||
| nPCR | nF | AACGGATTATTCTTTATAGCTTGCT | 55 | 546/565 | |||
| nR | GGCAGTATTAAAAGCAGCTCCAGG | ||||||
| B. burgdorferi s.l. | 23S rRNA | PCR | F | GCGAACGGGTGAGTAACG | 50 | 1360 | [39] |
| R | CCTCCCTTACGGGTTAGAA | ||||||
| 16S rRNA | PCR | F | GAGGCGAAGGCGAACTTCTG | 60.2 | 622 | [40] | |
| R | CTAGCGATTCCAACTTCATGAAG | ||||||
| C. burnetii | htpB | PCR | F | GCGGGTGATGGTACCACAACA | 57 | 501 | [41] |
| R | GGCAATCACCAATAAGGGCCG | ||||||
| nPCR | nF | TTGCTGGAATGAACCCCA | 52 | 325 | |||
| nR | TCAAGCTCCGCACTCATG | ||||||
| Rickettsia spp. | ompB | PCR | F | AAACAATAATCAAGGTACTGT | 55 | 212/209 | [42] |
| R | TACTTCCGGTTACAGCAAAGT | ||||||
| ompA | nPCR | nF | GCTTTATTCACCACCTCAAC | 811 | [43] | ||
| nR | TR(g/a)ATCACCACCGTAAGTAAAT | ||||||
| gltA | nPCR | nF | GCAAGTATCGGTGAGGATGTAAT | 401 | [44] | ||
| nR | GCTTCCTTAAAATTCAATAAATCAGGAT | ||||||
| Tick Species | Target Gene | GenBank Accession Number | Length (bp) | Identity (%) | Accession Number (Host, Country) |
|---|---|---|---|---|---|
| D. nuttalli | coxI | OQ816756 | 849 | 97.76% | OM368307 cattle China |
| H. qinghaiensis | coxI | OQ816757 | 849 | 97.76% | NC_062067 yak China |
| D. nuttalli | coxI | OQ816758 | 849 | 100% | MK028675 sheep China |
| H. qinghaiensis | coxI | OQ816759 | 849 | 99.18% | NC_062067 yak China |
| D. nuttalli | coxI | OQ816760 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816761 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816762 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816763 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816764 | 849 | 99.88% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816765 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816766 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816767 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816768 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816769 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816770 | 849 | 100% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816771 | 849 | 99.88% | MK028675 sheep China |
| D. nuttalli | coxI | OQ816772 | 849 | 100% | MK028675 sheep China |
| H. qinghaiensis | coxI | OQ816773 | 849 | 98.12% | OK094412 goat China |
| D. nuttalli | coxI | OQ816774 | 849 | 98.59% | OM368307 cattle China |
| H. qinghaiensis | coxI | OQ816775 | 849 | 98.23% | OK094412 goat China |
| H. qinghaiensis | coxI | OQ816776 | 849 | 98.12% | OK094412 goat China |
| H. qinghaiensis | coxI | OQ816777 | 849 | 98.82% | OK094412 goat China |
| D. nuttalli | coxI | OQ816778 | 849 | 97.53% | OM368307 cattle China |
| TBPs 1 | Sheep | goat | Cattle | Yak | Camel | Donkey | Horse | Tick | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| 52 | 67 | 49 | 43 | 50 | 40 | 65 | 62 | ||
| A. bovis | n. d. 2 | 44 (65.67; 12.02) 3 | 1 (2.04; 0.27) | n. d. | n. d. | n. d. | n. d. | 3 (4.83; 0.82) | 0.29 |
| A. capra | 8 (15.38; 2.19) | 10 (14.93; 2.73) | n. d. | n. d. | n. d. | n. d. | n. d. | 2 (3) | 0.29 |
| A. marginale | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | - |
| A. ovis | 24 (46.15; 6.56) | 1 (1.49; 0.27) | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | 0.31 |
| A. phagocytophilum | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | - |
| B. burgdorferi s.l. | 14 (26.92; 3.83) | 13 (19.40; 3.55) | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | 0.31 |
| C. burnetii | 2 (3.84; 0.55) | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | 1 (1.61; 0.27) | 0.31 |
| Rickettsia spp. | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | n. d. | 2 (3.23; 0.55) | 0.33 |
| Obtained Sequences | The Closest BLASTn Match | ||||||
|---|---|---|---|---|---|---|---|
| Pathogen | Animal | Target Gene | GenBank Accession Number | Length (bp) | Identity (%) | Pathogen Isolate | Accession Number (Host, Country) |
| A. bovis | cattle | 16s rRNA | OQ826693 | 551 | 99.64% | Uncultured Anaplasma sp. | KM186933 dzeren China |
| goat | 16s rRNA | OQ826694 | 551 | 99.82% | Uncultured Anaplasma sp. | KM186933 dzeren China | |
| goat | 16s rRNA | OQ826695 | 551 | 99.64% | Uncultured Anaplasma sp. | KM186933 dzeren China | |
| goat | 16s rRNA | OQ826696 | 548 | 100% | A. bovis | MN213735 giraffe Pakistan | |
| goat | 16s rRNA | OQ826697 | 551 | 99.82% | Uncultured Anaplasma sp. | MZ069112 yak China | |
| goat | 16s rRNA | OQ826698 | 551 | 99.82% | Uncultured Anaplasma sp. | KM186933 dzeren China | |
| goat | 16s rRNA | OQ826699 | 551 | 99.82% | Uncultured Anaplasma sp. | KM186933 dzeren China | |
| goat | 16s rRNA | OQ826700 | 551 | 100% | A. bovis | MN213735 giraffe Pakistan | |
| goat | 16s rRNA | OQ826701 | 551 | 100% | A. bovis | MN213735 giraffe Pakistan | |
| goat | 16s rRNA | OQ826702 | 551 | 99.82% | Uncultured Anaplasma sp. | MZ069112 yak China | |
| tick | 16s rRNA | OQ826703 | 551 | 100% | Uncultured Anaplasma sp. | MZ069112 yak China | |
| tick | 16s rRNA | OQ826704 | 551 | 99.27% | Uncultured Anaplasma sp. | KM186933 dzeren China | |
| tick | 16s rRNA | OQ826705 | 551 | 99.46% | A. bovis | MN213735 giraffe Pakistan | |
| A. ovis | goat | msp4 | OQ847093 | 347 | 100% | A. ovis | MN394791 Tibetan sheep China |
| Tibetan sheep | msp4 | OQ847094 | 347 | 99.71% | A. ovis | MN394791 Tibetan sheep China | |
| A. capra | tick | gltA | OQ847092 | 793 | 99.37% | A. capra | MZ130266 Tibetan sheep China |
| goat | gltA | OQ847085 | 793 | 98.63% | A. capra | MW930535 goat France | |
| goat | gltA | OQ847086 | 793 | 98.49% | A. capra | MW930535 goat France | |
| Tibetan sheep | gltA | OQ847088 | 793 | 99.75% | A. capra | MZ130266 Tibetan sheep China | |
| Tibetan sheep | gltA | OQ847089 | 793 | 99.62% | A. capra | MZ130266 Tibetan sheep China | |
| Tibetan sheep | gltA | OQ847090 | 793 | 99.62% | A. capra | MZ130264 Tibetan sheep China | |
| Tibetan sheep | gltA | OQ847091 | 793 | 97.86% | A. capra | MZ130266 Tibetan sheep China | |
| Rickettsia spp. | tick | gltA | OQ872771 | 400 | 100% | R. raoultii | MK304547 tick Russia |
| tick | gltA | OQ872772 | 400 | 100% | R. raoultii | MT178338 tick China | |
| Borrelia spp. | goat | 16s rRNA | OQ875206 | 623 | 99.52% | Borrelia sp. | KY284014 camel China |
| Tibetan sheep | 16s rRNA | OQ875207 | 623 | 99.84% | Borrelia sp. | KY284014 camel China | |
| C. burnetii | tick | htpB | OQ872773 | 325 | 99.69% | C. burnetii | MK416231 tick Tunisia |
| Tibetan sheep | htpB | OQ872774 | 325 | 99.69% | C. burnetii | MK416231 tick Tunisia | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Jian, Y.; Wang, G.; Zafar, I.; Li, X.; Wang, G.; Hu, Y.; Yokoyama, N.; Ma, L.; Xuan, X. Epidemiological Investigation of Tick-Borne Bacterial Pathogens in Domestic Animals from the Qinghai–Tibetan Plateau Area, China. Pathogens 2024, 13, 86. https://doi.org/10.3390/pathogens13010086
Ma Y, Jian Y, Wang G, Zafar I, Li X, Wang G, Hu Y, Yokoyama N, Ma L, Xuan X. Epidemiological Investigation of Tick-Borne Bacterial Pathogens in Domestic Animals from the Qinghai–Tibetan Plateau Area, China. Pathogens. 2024; 13(1):86. https://doi.org/10.3390/pathogens13010086
Chicago/Turabian StyleMa, Yihong, Yingna Jian, Geping Wang, Iqra Zafar, Xiuping Li, Guanghua Wang, Yong Hu, Naoaki Yokoyama, Liqing Ma, and Xuenan Xuan. 2024. "Epidemiological Investigation of Tick-Borne Bacterial Pathogens in Domestic Animals from the Qinghai–Tibetan Plateau Area, China" Pathogens 13, no. 1: 86. https://doi.org/10.3390/pathogens13010086
APA StyleMa, Y., Jian, Y., Wang, G., Zafar, I., Li, X., Wang, G., Hu, Y., Yokoyama, N., Ma, L., & Xuan, X. (2024). Epidemiological Investigation of Tick-Borne Bacterial Pathogens in Domestic Animals from the Qinghai–Tibetan Plateau Area, China. Pathogens, 13(1), 86. https://doi.org/10.3390/pathogens13010086








