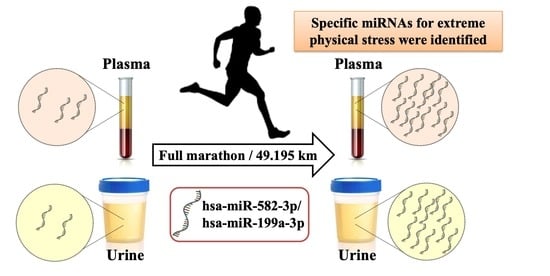
A Pilot Study of miRNA Expression Profile as a Liquid Biopsy for Full-Marathon Participants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval and Study Overview
2.2. Study Participants
2.3. Sample Collection
2.4. Extracted Exosomal RNA in Plasma and Urine
2.5. Library Preparation for Small RNA Sequencing
2.6. Data Analysis Process
2.7. Selection of miRNA Potential Biomarkers by Filtering with Sufficient Expression and Fold Change
3. Results
3.1. Quality Information of the NGS Run and Informatics Analysis
3.2. MiRNA Expression Profile during Full Marathon
3.3. Timeline of Known Muscle-Specific MiRNA Expression Patterns in Plasma and Urine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olympic Marathon: A Centennial History of the Games’ Most Storied Race in: Sport History Review. 2001. Available online: https://journals.humankinetics.com/view/journals/shr/32/1/article-p62.xml (accessed on 20 June 2021).
- Lee, E.C.; Fragala, M.S.; Kavouras, S.; Queen, R.M.; Pryor, J.L.; Casa, D.J. Biomarkers in Sports and Exercise: Tracking Health, Performance, and Recovery in Athletes. J. Strength Cond. Res. 2017, 31, 2920–2937. [Google Scholar] [CrossRef] [Green Version]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufresne, S.; Rébillard, A.; Muti, P.; Friedenreich, C.; Brenner, D.R. A Review of Physical Activity and Circulating miRNA Expression: Implications in Cancer Risk and Progression. Cancer Epidemiol. Biomark. Prev. 2017, 27, 11–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidrich, I.; Ačkar, L.; Mohammadi, P.M.; Pantel, K. Liquid biopsies: Potential and challenges. Int. J. Cancer 2021, 148, 528–545. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Qu, X. Cancer biomarker detection: Recent achievements and challenges. Chem. Soc. Rev. 2015, 44, 2963–2997. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Allegra, A.; Alonci, A.; Campo, S.; Penna, G.; Petrungaro, A.; Gerace, D.; Musolino, C. Circulating microRNAs: New biomarkers in diagnosis, prognosis and treatment of cancer (Review). Int. J. Oncol. 2012, 41, 1897–1912. [Google Scholar] [CrossRef] [Green Version]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Lv, Y.; Li, G.; Xiao, J. MicroRNAs in heart and circulation during physical exercise. J. Sport Health Sci. 2018, 7, 433–441. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Dávalos, A.; Montero, A.; García-González, Á.; Tyshkovska, I.; González-Medina, A.; Soares, S.M.A.; Martínez-Camblor, P.; Casas-Agustench, P.; Rabadán, M.; et al. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J. Appl. Physiol. 2015, 119, 124–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Max, K.E.A.; Bertram, K.; Akat, K.M.; Bogardus, K.A.; Li, J.; Morozov, P.; Ben-Dov, I.Z.; Li, X.; Weiss, Z.; Azizian, A.; et al. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc. Natl. Acad. Sci. 2018, 115, E5334–E5343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siracusa, J.; Koulmann, N.; Banzet, S. Circulating myomiRs: A new class of biomarkers to monitor skeletal muscle in physiology and medicine. J. Cachexia Sarcopenia Muscle 2017, 9, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Mall, C.; Rocke, D.M.; Durbin-Johnson, B.; Weiss, R.H. Stability of miRNA in human urine supports its biomarker potential. Biomark. Med. 2013, 7, 623–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lässer, C. Exosomal RNA as biomarkers and the therapeutic potential of exosome vectors. Expert Opin. Biol. Ther. 2012, 12, S189–S197. [Google Scholar] [CrossRef]
- Estébanez, B.; Jiménez-Pavón, D.; Huang, C.; Cuevas, M.J.; González-Gallego, J. Effects of exercise on exosome release and cargo in in vivo and ex vivo models: A systematic review. J. Cell. Physiol. 2021, 236, 3336–3353. [Google Scholar] [CrossRef]
- Sugasawa, T.; Fujita, S.-I.; Kuji, T.; Ishibashi, N.; Tamai, K.; Kawakami, Y.; Takekoshi, K. Dynamics of Specific cfDNA Fragments in the Plasma of Full Marathon Participants. Genes 2021, 12, 676. [Google Scholar] [CrossRef]
- Soplinska, A.; Zareba, L.; Wicik, Z.; Eyileten, C.; Jakubik, D.; Siller-Matula, J.M.; De Rosa, S.; Malek, L.A.; Postula, M. MicroRNAs as Biomarkers of Systemic Changes in Response to Endurance Exercise—A Comprehensive Review. Diagnostics 2020, 10, 813. [Google Scholar] [CrossRef]
- Shishikura, Y.; Tokinoya, K.; Aita, Y.; Sekine, N.; Sugasawa, T.; Yoshida, Y.; Kosaki, K.; Kumamoto, S.; Ishikura, K.; Kuji, T.; et al. The Dynamics of Cell-Free DNA from Urine and Blood after a Full Marathon. bioRxiv 2021. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Friedlander, M.; Mackowiak, S.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; et al. Integrated Profiling of MicroRNAs and mRNAs: MicroRNAs Located on Xq27.3 Associate with Clear Cell Renal Cell Carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2009, 26, 136–138. [Google Scholar] [CrossRef]
- Storey, J. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Metpally, R.P.R.; Nasser, S.; Malenica, I.; Courtright, A.; Carlson, E.; Ghaffari, L.; Villa, S.; Tembe, W.; Van Keuren-Jensen, K. Comparison of Analysis Tools for miRNA High Throughput Sequencing Using Nerve Crush as a Model. Front. Genet. 2013, 4, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.; Hu, G.; He, X.; Zhou, P.; Li, J.; He, B.; Sun, W. MicroRNA-424-5p Suppresses the Expression of SOCS6 in Pancreatic Cancer. Pathol. Oncol. Res. 2013, 19, 739–748. [Google Scholar] [CrossRef]
- Connolly, M.; Paul, R.; Garros, R.F.; Natanek, S.A.; Bloch, S.; Lee, J.; Lorenzo, J.P.; Patel, H.; Cooper, C.; Sayer, A.A.; et al. miR-424-5p reduces ribosomal RNA and protein synthesis in muscle wasting. J. Cachexia Sarcopenia Muscle 2018, 9, 400–416. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.W.; Sun, X.; Yu, Y.; Zhao, H.M.; Yang, Z.J.; Wang, X.; Cao, X.C. miR-361-5p suppresses lung cancer cell lines progression by targeting FOXM1. Neoplasma 2017, 64, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shao, R.; Gao, W.; Sun, G.; Liu, Y.; Fa, X. Inhibition of miR-361-5p suppressed pulmonary artery smooth muscle cell survival and migration by targeting ABCA1 and inhibiting the JAK2/STAT3 pathway. Exp. Cell Res. 2018, 363, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Chai, B.; Guo, Y.; Cui, X.; Liu, J.; Suo, Y.; Dou, Z.; Li, N. MiR-223-3p promotes the proliferation, invasion and migration of colon cancer cells by negative regulating PRDM1. Am. J. Transl. Res. 2019, 11, 4516–4523. [Google Scholar] [PubMed]
- Cheng, N.; Liu, C.; Li, Y.; Gao, S.; Han, Y.-C.; Wang, X.; Du, J.; Zhang, C. MicroRNA-223-3p promotes skeletal muscle regeneration by regulating inflammation in mice. J. Biol. Chem. 2020, 295, 10212–10223. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Peng, J.; He, S.; Huang, H.; Lin, L.; Zhu, Q.; Ye, L.; Li, T.; Zhang, X.; Gao, Y.; et al. miR-223-5p targeting ERG inhibits prostate cancer cell proliferation and migration. J. Cancer 2020, 11, 4453–4463. [Google Scholar] [CrossRef]
- de Melo Maia, B.; Rodrigues, I.S.; Akagi, E.M.; Soares do Amaral, N.; Ling, H.; Monroig, P.; Soares, F.A.; Calin, G.A.; Rocha, R.M. MiR-223-5p works as an oncomiR in vulvar carcinoma by TP63 suppression. Oncotarget 2016, 7, 49217–49231. [Google Scholar] [CrossRef] [Green Version]
- Dou, L.; Han, K.; Xiao, M.; Lv, F. miR-223-5p Suppresses Tumor Growth and Metastasis in Non-Small Cell Lung Cancer by Targeting E2F8. Oncol. Res. 2019, 27, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, S.; Yamada, Y.; Arai, T.; Okato, A.; Idichi, T.; Kato, M.; Koshizuka, K.; Ichikawa, T.; Seki, N. Dual strands of the miR-223 duplex (miR-223-5p and miR-223-3p) inhibit cancer cell aggressiveness: Targeted genes are involved in bladder cancer pathogenesis. J. Hum. Genet. 2018, 63, 657–668. [Google Scholar] [CrossRef]
- Conickx, G.; Mestdagh, P.; Cobos, F.A.; Verhamme, F.M.; Maes, T.; Vanaudenaerde, B.M.; Seys, L.J.M.; LaHousse, L.; Kim, R.Y.; Hsu, A.C.; et al. MicroRNA Profiling Reveals a Role for MicroRNA-218-5p in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2017, 195, 43–56. [Google Scholar] [CrossRef]
- Huang, L.; Ding, Y.; Yang, L.; Jiang, X.; Xia, Z.; You, Z. The effect of LncRNA SNHG16 on vascular smooth muscle cells in CHD by targeting miRNA-218-5p. Exp. Mol. Pathol. 2021, 118, 104595. [Google Scholar] [CrossRef]
- MiR-517a-3p Accelerates Lung Cancer Cell Proliferation and Invasion through Inhibiting FOXJ3 Expression—ScienceDirect. Available online: https://www.sciencedirect.com/science/article/abs/pii/S0024320514004792?casa_token=2jxHhVmesZgAAAAA:i5_hLykCnWi1H8NjuhObl1BeGMUynmOzDh8zIIsKhpr8E6mNufbK6D44OvrRPpyWE4Zfe3a6rN8 (accessed on 29 May 2021).
- Fernández-Sanjurjo, M.; Úbeda, N.; Fernández-García, B.; Del Valle, M.; De Molina, A.R.; Crespo, M.C.; Martín-Hernández, R.; Casas-Agustench, P.; Martínez-Camblor, P.; De Gonzalo-Calvo, D.; et al. Exercise dose affects the circulating microRNA profile in response to acute endurance exercise in male amateur runners. Scand. J. Med. Sci. Sports 2020, 30, 1896–1907. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.; Oliveira, G.; Madrid, B.; Almeida, J.A.; Franco, O.; Pereira, R. Circulating miR-1, miR-133a, and miR-206 levels are increased after a half-marathon run. Biomarkers 2014, 19, 585–589. [Google Scholar] [CrossRef]
- D’Souza, R.F.; Markworth, J.F.; Aasen, K.M.M.; Zeng, N.; Cameron-Smith, D.; Mitchell, C.J. Acute resistance exercise modulates microRNA expression profiles: Combined tissue and circulatory targeted analyses. PLoS ONE 2017, 12, e0181594. [Google Scholar] [CrossRef] [PubMed]
- Camera, D.M.; Ong, J.N.; Coffey, V.G.; Hawley, J.A. Selective Modulation of MicroRNA Expression with Protein Ingestion Following Concurrent Resistance and Endurance Exercise in Human Skeletal Muscle. Front. Physiol. 2016, 7, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, L.; Cai, J.; Chen, B.; Wu, S.; Li, R.; Baixue, C.; Yang, Y.; Guan, H.; Zhu, X.; Zhang, L.; et al. Aberrantly expressed miR-582-3p maintains lung cancer stem cell-like traits by activating Wnt/β-catenin signalling. Nat. Commun. 2015, 6, 8640. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Lu, S.; Dong, L.; Xue, Y.; Yao, C.; Tong, C.; Wang, C.; Shu, X. hsa-miR-320d and hsa-miR-582, miRNA Biomarkers of Aortic Dissection, Regulate Apoptosis of Vascular Smooth Muscle Cells. J. Cardiovasc. Pharmacol. 2018, 71, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, R.; Nishimura, J.; Kagawa, Y.; Osawa, H.; Hasegawa, J.; Murata, K.; Okamura, S.; Ota, H.; Uemura, M.; Hata, T.; et al. Circulating miR-199a-3p as a novel serum biomarker for colorectal cancer. Oncol. Rep. 2014, 32, 2354–2358. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Li, Y.-F.; Wu, G.-F.; Song, Z.-Y.; Lu, H.-Z.; Song, C.-C.; Zhang, Q.-L.; Zhu, J.-Y.; Yang, G.-S.; Shi, X.-E. MiRNA-199a-3p Regulates C2C12 Myoblast Differentiation through IGF-1/AKT/mTOR Signal Pathway. Int. J. Mol. Sci. 2013, 15, 296–308. [Google Scholar] [CrossRef] [Green Version]
- Jacques, M.; Hiam, D.; Craig, J.; Barrès, R.; Eynon, N.; Voisin, S. Epigenetic changes in healthy human skeletal muscle following exercise–A systematic review. Epigenetics 2019, 14, 633–648. [Google Scholar] [CrossRef]
- Clauss, S.; Wakili, R.; Hildebrand, B.; Kaab, S.; Hoster, E.; Klier, I.; Martens, E.; Hanley, A.; Hanssen, H.; Halle, M.; et al. MicroRNAs as Biomarkers for Acute Atrial Remodeling in Marathon Runners (The miRathon Study—A Sub-Study of the Munich Marathon Study). PLoS ONE 2016, 11, e0148599. [Google Scholar] [CrossRef]
- Baggish, A.L.; Park, J.; Min, P.-K.; Isaacs, S.; Parker, B.A.; Thompson, P.D.; Troyanos, C.; D’Hemecourt, P.; Dyer, S.; Thiel, M.; et al. Rapid upregulation and clearance of distinct circulating microRNAs after prolonged aerobic exercise. J. Appl. Physiol. 2014, 116, 522–531. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.J.; Roberts, D. Effects of high volume and/or intense exercise on selected blood chemistry parameters. Clin. Biochem. 1994, 27, 435–440. [Google Scholar] [CrossRef]
- Priest, M.J.B.; Oei, M.T.O.; Moorehead, P.W.R. Exercise-induced Changes in Common Laboratory Tests. Am. J. Clin. Pathol. 1982, 77, 285–289. [Google Scholar] [CrossRef] [Green Version]
- Arneth, B. Update on the types and usage of liquid biopsies in the clinical setting: A systematic review. BMC Cancer 2018, 18, 527. [Google Scholar] [CrossRef] [Green Version]
- Jeffries, J.; Zhou, W.; Hsu, A.; Deng, Q. miRNA-223 at the crossroads of inflammation and cancer. Cancer Lett. 2019, 451, 136–141. [Google Scholar] [CrossRef]
- Haneklaus, M.; Gerlic, M.; O’Neill, L.A.J.; Masters, S.L. miR-223: Infection, inflammation and cancer. J. Intern. Med. 2013, 274, 215–226. [Google Scholar] [CrossRef]
- Aziz, F. The emerging role of miR-223 as novel potential diagnostic and therapeutic target for inflammatory disorders. Cell. Immunol. 2016, 303, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Taïbi, F.; Meuth, V.M.-L.; Massy, Z.A.; Metzinger, L. miR-223: An inflammatory oncomiR enters the cardiovascular field. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2014, 1842, 1001–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, A.P.; Lamon, S.; Boon, H.; Wada, S.; Güller, I.; Brown, E.L.; Chibalin, A.; Zierath, J.; Snow, R.J.; Stepto, N.; et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 2013, 591, 4637–4653. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, X.; Zheng, Y.; Li, F.; Lu, Z.; Chen, C.; Liu, J.; Wang, Y.; Peng, Y.; Shen, Z.; et al. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp. Cell Res. 2012, 318, 2324–2334. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Pan, X.; Li, Y.; Hu, Y.; Tao, L.; Li, Z.; Zhao, L.; Wang, J.; Li, H.; Lai, Y.; et al. MiR-23a-3p acts as an oncogene and potential prognostic biomarker by targeting PNRC2 in RCC. Biomed. Pharmacother. 2019, 110, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Bemben, M.G.; Bemben, D.A. Bone and muscle specific circulating microRNAs in postmenopausal women based on osteoporosis and sarcopenia status. Bone 2019, 120, 271–278. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baggish, A.L.; Hale, A.; Weiner, R.B.; Lewis, G.D.; Systrom, D.; Wang, F.; Wang, T.; Chan, S.Y. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 2011, 589, 3983–3994. [Google Scholar] [CrossRef] [PubMed]






| Type of RNA | Example of Characteristic | Sample with Expression | Reference |
|---|---|---|---|
| hsa-miR-424-5p | Pancreatic cancer, muscle, and inflammation | Plasma | [12,28,29] |
| hsa-miR-361-5p | Lung cancer and muscle (smooth muscle) | Plasma | [30,31] |
| hsa-miR-223-3p | Colon cancer, muscle, and inflammation | Plasma | [12,32,33] |
| hsa-miR-223-5p | Malignant neoplasms including vulvar carcinoma, non-small cell lung cancer, bladder cancer, prostate cancer, and inflammation | Plasma | [12,34,35,36,37] |
| hsa-miR-218-5p | Chronic obstructive pulmonary disease and muscle (smooth muscle) | Urine | [38,39] |
| hsa-miR-3158-3p | Unknown | Urine | |
| hsa-miR-3158-5p | Unknown | Urine | |
| hsa-miR-517a-3p | Lung cancer and inflammation | Urine | [12,40] |
| hsa-miR-1-3p | Muscle specific | Plasma and Urine | [4,19,41] |
| hsa-miR-206 | Muscle specific | Plasma and Urine | [4,19,42] |
| hsa-miR-23a-3p | Muscle | Plasma and Urine | [43,44] |
| hsa-miR-582-3p | Lung cancer and muscle (smooth muscle) | Plasma and Urine | [45,46] |
| hsa-miR-199a-3p | Colorectal cancer and muscle | Plasma and Urine | [47,48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuji, T.; Sugasawa, T.; Fujita, S.-i.; Ono, S.; Kawakami, Y.; Takekoshi, K. A Pilot Study of miRNA Expression Profile as a Liquid Biopsy for Full-Marathon Participants. Sports 2021, 9, 134. https://doi.org/10.3390/sports9100134
Kuji T, Sugasawa T, Fujita S-i, Ono S, Kawakami Y, Takekoshi K. A Pilot Study of miRNA Expression Profile as a Liquid Biopsy for Full-Marathon Participants. Sports. 2021; 9(10):134. https://doi.org/10.3390/sports9100134
Chicago/Turabian StyleKuji, Tomoaki, Takehito Sugasawa, Shin-ichiro Fujita, Seiko Ono, Yasushi Kawakami, and Kazuhiro Takekoshi. 2021. "A Pilot Study of miRNA Expression Profile as a Liquid Biopsy for Full-Marathon Participants" Sports 9, no. 10: 134. https://doi.org/10.3390/sports9100134