Mamestra brassicae Multiple Nucleopolyhedroviruses Prevents Pupation of Helicoverpa armigera by Regulating Juvenile Hormone Titer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects and Viruses
2.2. Virus Infection and Juvenile Hormone Determination
2.3. RNA Extraction and Quality Control
2.4. Library Preparation, mRNAs Sequencing, and Analysis
2.5. Validation of mRNA by Real-Time Quantitative PCR
2.6. The Expression Patterns, Synthesis of dsRNA, RNA Interference
2.7. Viral Infection
2.8. Data Analysis
3. Results
3.1. MbMNPV Infection Prolonged the Larval Stage and Caused the Increase of JH Titer
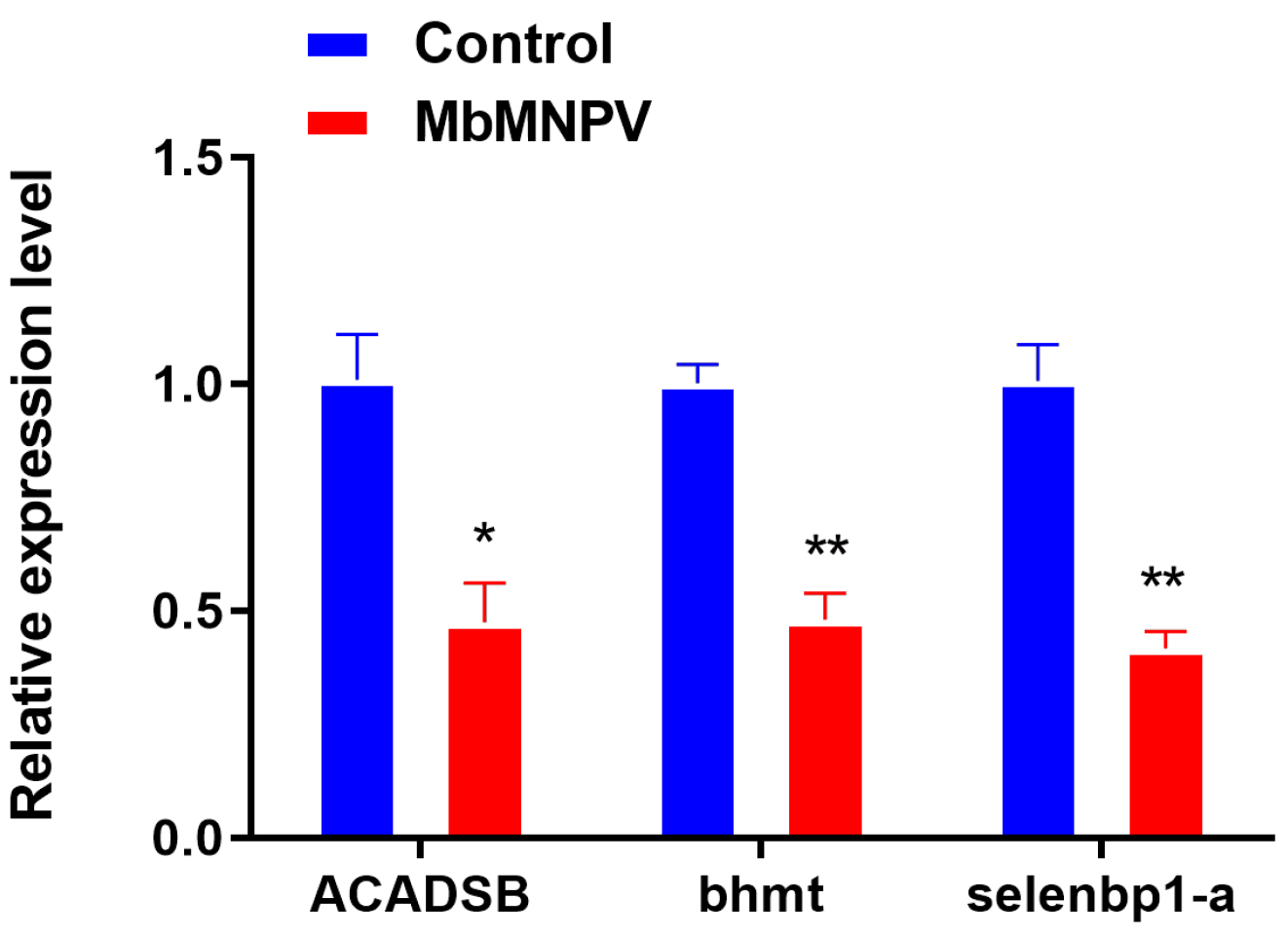
3.2. RNA-Seq Sequencing and Data Analysis
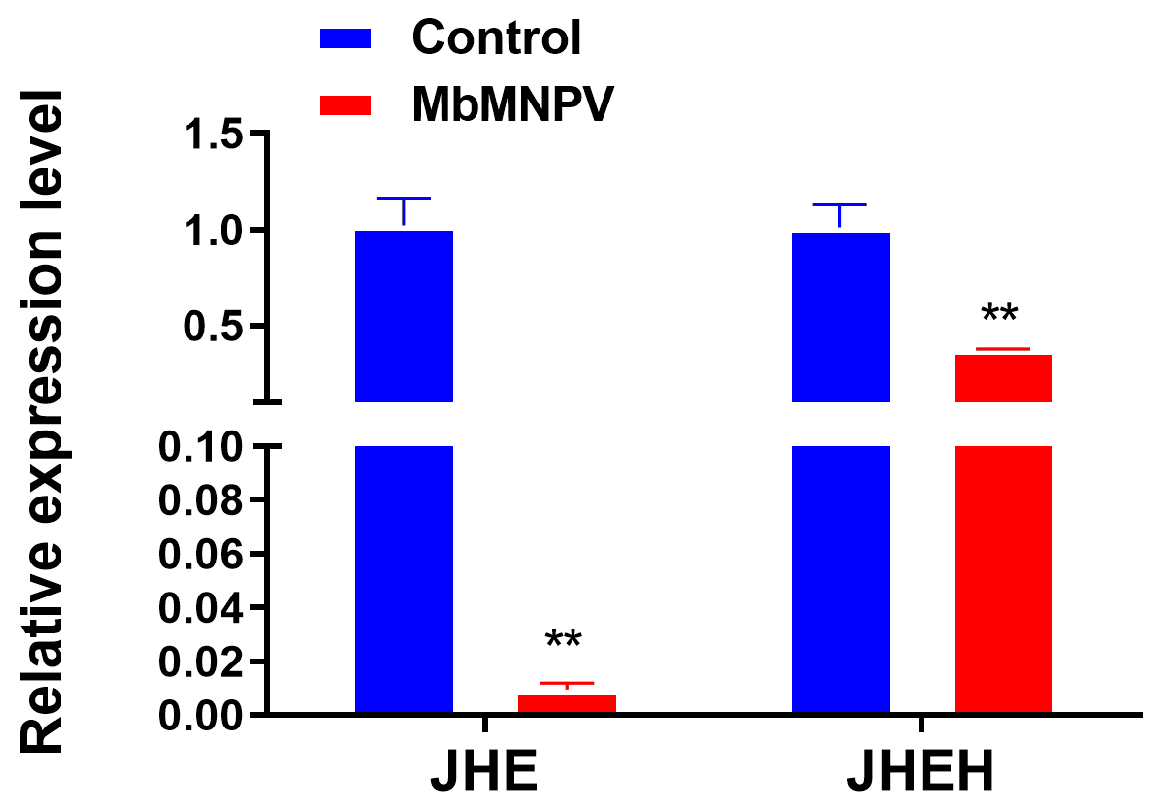
3.3. MbMNPV Infection Inhibited the Expression of JHE and JHEH
3.4. Effects of Knocking down of HaJHE and HaJHEH on the Pupation of H. armigera
3.5. Detection of Expression Patterns of JHE and JHEH
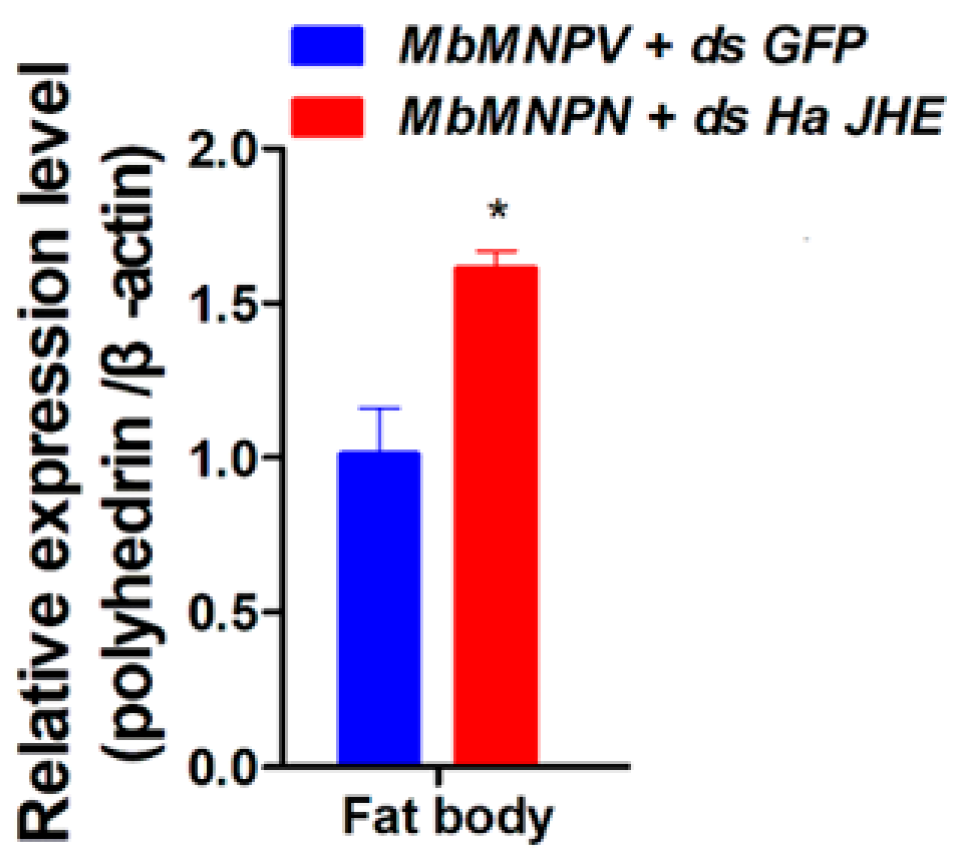
3.6. Knockdown of HaJHE Promoted the MbMNPV Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ahmad, M.; Rasool, B.; Ahmad, M.; Russell, D.A. Resistance and Synergism of Novel Insecticides in Field Populations of Cotton Bollworm Helicoverpa armigera (Lepidoptera: Noctuidae) in Pakistan. J. Econ. Entomol. 2019, 112, 859–871. [Google Scholar] [CrossRef]
- Dong, W.; Qiu, X.; Ren, X.; Fang, N.; Wang, K. Resistance selection and biochemical characterization of spinosad resistance in Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 2009, 95, 90–94. [Google Scholar]
- Palli, S.R.; Retnakaran, A. Biological control of forest pests: A biotechnological perspective. In Forest Products Biotechnology; CRC Press: Boca Raton, FL, USA, 1997; pp. 277–296. [Google Scholar]
- Doyle, C.J.; Hirst, M.L.; Cory, J.S.; Entwistle, P.F. Risk Assessment Studies: Detailed Host Range Testing of Wild-Type Cabbage Moth, Mamestra brassicae (Lepidoptera: Noctuidae), Nuclear Polyhedrosis Virus. Appl. Environ. Microbiol. 1990, 56, 2704–2710. [Google Scholar] [CrossRef] [PubMed]
- Hostetter, D.L.; Puttler, B. A New Broad Host Spectrum Nuclear Polyhedrosis Virus Isolated from a Celery Looper, Anagrapha falcifera (Kirby), (Lepidoptera: Noctuidae). Environ. Entomol. 1991, 20, 1480–1488. [Google Scholar] [CrossRef]
- Ayres, M.D.; Howard, S.C.; Kuzio, J.; Lopez-Ferber, M.; Possee, R.D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 1994, 202, 586–605. [Google Scholar] [CrossRef]
- Harrison, R.L.; Bonning, B.C. The nucleopolyhedroviruses of Rachiplusia ou and Anagrapha falcifera are isolates of the same virus. J. Gen. Virol. 1999, 80 Pt 10, 2793–2798. [Google Scholar] [CrossRef]
- Lawrence, P.O.; Lanzrein, B. Hormonal Interactions between Insect Endoparasites and Their Host Insects—ScienceDirect. Parasites Pathog. Insects 1993, 59–86. [Google Scholar] [CrossRef]
- Edwards, J.P.; Weaver, R.J. Endocrine changes in lepidopteran larvae—Potential challenges to parasitoid development and survival. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2000, 126, S31. [Google Scholar] [CrossRef]
- Kiuchi, M.; Yasui, H.; Hayasaka, S.; Kamimura, M. Entomogenous fungus Nomuraea rileyi inhibits host insect molting by C22-oxidizing inactivation of hemolymph ecdysteroids. Arch. Insect Biochem. Physiol. 2003, 52, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Nakai, M.; Shiotsuki, T.; Kunimi, Y. An entomopoxvirus and a granulovirus use different mechanisms to prevent pupation of Adoxophyes honmai. Virus Res. 2004, 101, 185–191. [Google Scholar] [CrossRef]
- O’reilly, D.R. Baculovirus-encoded ecdysteroid UDP- glucosyl-transferase. Insect Biochem. Mol. Biol. 1995, 25, 541–550. [Google Scholar] [CrossRef]
- O’reilly, D.R.; Miller, L.K. A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyl transferase. Science 1989, 245, 1110–1112. [Google Scholar] [CrossRef]
- Wyatt, G.; Davey, K. Cellular and molecular actions of juvenile hormone. II. Roles of juvenile hormones in adult insects. Adv. Insect Physiol. 1996, 26, 1–55. [Google Scholar]
- Kamita, S.G.; Hammock, B.D. Juvenile hormone esterase: Biochemistry and structure. J. Pestic. Sci. 2010, 35, 265–274. [Google Scholar] [CrossRef]
- Tsubota, T.; Nakakura, T.; Shiotsuki, T. Molecular characterization and enzymatic analysis of juvenile hormone epoxide hydrolase genes in the red flour beetle Tribolium castaneum. Insect Mol. Biol. 2010, 19, 399–408. [Google Scholar] [CrossRef]
- Flatt, T.; Kawecki, T.J. Pleiotropic effects of methoprene-tolerant (Met), a gene involved in juvenile hormone metabolism, on life history traits in Drosophila melanogaster. Genetica 2004, 122, 141–160. [Google Scholar] [CrossRef]
- Kayukawa, T.; Minakuchi, C.; Namiki, T.; Togawa, T.; Yoshiyama, M.; Kamimura, M.; Mita, K.; Imanishi, S.; Kiuchi, M.; Ishikawa, Y.; et al. Transcriptional regulation of juvenile hormone-mediated induction of Krüppel homolog 1, a repressor of insect metamorphosis. Proc. Natl. Acad. Sci. USA 2012, 109, 11729–11734. [Google Scholar] [CrossRef]
- Wyatt, G.R. Comprehensive insect physiology, biochemistry, and pharmacology. Int. J. Biochem. 1985, 17, 1281–1282. [Google Scholar] [CrossRef]
- Ismail, S.; Goin, C.; Muthumani, K.; Kim, M.; Dahm, K.; Bhaskaran, G. Juvenile hormone acid and ecdysteroid together induce competence for metamorphosis of the Verson’s gland in Manduca sexta. J. Insect Physiol. 2000, 46, 59–68. [Google Scholar] [CrossRef]
- Ismail, S.M.; Satyanarayana, K.; Bradfield, J.Y.; Dahm, K.H.; Bhaskaran, G. Juvenile hormone acid: Evidence for a hormonal function in induction of vitellogenin in larvae of Manduca sexta. Arch. Insect Biochem. Physiol. 1998, 37, 305–314. [Google Scholar] [CrossRef]
- Minakuchi, C.; Namiki, T.; Shinoda, T. Krüppel homolog 1, an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev. Biol. 2009, 325, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Abdou, M.A.; He, Q.; Wen, D.; Zyaan, O.; Wang, J.; Xu, J.; Baumann, A.A.; Joseph, J.; Wilson, T.G.; Li, S.; et al. Drosophila Met and Gce are partially redundant in transducing juvenile hormone action. Insect Biochem. Mol. Biol. 2011, 41, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Skowronek, P.; Wójcik, Ł.; Strachecka, A. Fat Body-Multifunctional Insect Tissue. Insects 2021, 12, 547. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.C.; Ma, B.W.; Berg, B.G.; Xie, G.Y.; Tang, Q.B.; Guo, X.R. A global-wide search for sexual dimorphism of glomeruli in the antennal lobe of female and male Helicoverpa armigera. Sci. Rep. 2016, 6, 35204. [Google Scholar] [CrossRef] [PubMed]
- Kyeiccoku, G.K.; Kunimi, Y. Effect of entomopoxvirus infection of Pseudaletia separata larvae on the oviposition behavior of Cotesia kariyai. Entomol. Exp. Appl. 2010, 83, 93–97. [Google Scholar]
- Yao, S.; Yang, Y.; Xue, Y.; Zhao, W.; Liu, X.; Du, M.; Yin, X.; Guan, R.; Wei, J.; An, S. New insights on the effects of spinosad on the development of Helicoverpa armigera. Ecotoxicol. Environ. Saf. 2021, 221, 112452. [Google Scholar] [CrossRef]
- Wang, S.A.-O.; Wang, L.L.; Pu, Y.X.; Liu, J.Y.; Wang, M.X.; Zhu, J.; Shen, Z.Y.; Shen, X.J.; Tang, S.A.-O. Exorista sorbillans (Diptera: Tachinidae) parasitism shortens host larvae growth duration by regulating ecdysone and juvenile hormone titers in Bombyx mori (Lepidoptera: Bombycidae). J. Insect Sci. 2023, 23, 6. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Truman, J.W.; Riddiford, L.M. Endocrine insights into the evolution of metamorphosis in insects. Annu. Rev. Entomol. 2002, 47, 467–500. [Google Scholar] [CrossRef] [PubMed]
- Nakai, M.; Kinjo, H.; Takatsuka, J.; Shiotsuki, T.; Kamita, S.G.; Kunimi, Y. Entomopoxvirus infection induces changes in both juvenile hormone and ecdysteroid levels in larval Mythimna separata. J. Gen. Virol. 2016, 97, 225–232. [Google Scholar] [CrossRef]
- Palli, S.R.; Ladd, T.R.; Tomkins, W.L.; Shu, S.; Ramaswamy, S.B.; Tanaka, Y.; Arif, B.; Retnakaran, A. Choristoneura fumiferana entomopoxvirus prevents metamorphosis and modulates juvenile hormone and ecdysteroid titers. Insect Biochem. Mol. Biol. 2000, 30, 869–876. [Google Scholar] [CrossRef]
- Tremblay, N.; Baril, M.; Chatel-Chaix, L.; Es-Saad, S.; Park, A.Y.; Koenekoop, R.K.; Lamarre, D. Spliceosome SNRNP200 Promotes Viral RNA Sensing and IRF3 Activation of Antiviral Response. PLoS Pathog. 2016, 12, e1005772. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Mohr, I. Ribosome biogenesis restricts innate immune responses to virus infection and DNA. eLife 2019, 8, e49551. [Google Scholar] [CrossRef]
- Lee, E.Y.; Kim, S.; Kim, M.H. Aminoacyl-tRNA synthetases, therapeutic targets for infectious diseases. Biochem. Pharmacol. 2018, 154, 424–434. [Google Scholar] [CrossRef]
- Abernathy, E.; Glaunsinger, B. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 2015, 479–480, 600–608. [Google Scholar] [CrossRef]
- De Maio, F.A.; Risso, G.; Iglesias, N.G.; Shah, P.; Pozzi, B.; Gebhard, L.G.; Mammi, P.; Mancini, E.; Yanovsky, M.J.; Andino, R.; et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog. 2016, 12, e1005841. [Google Scholar] [CrossRef]
- Shukla, S.K.; Kumar, V. Hepatitis B virus X protein and c-Myc cooperate in the upregulation of ribosome biogenesis and in cellular transformation. FEBS J. 2012, 279, 3859–3871. [Google Scholar] [CrossRef] [PubMed]
- Duchon, A.A.; St Gelais, C.; Titkemeier, N.; Hatterschide, J.; Wu, L.; Musier-Forsyth, K. HIV-1 Exploits a Dynamic Multi-aminoacyl-tRNA Synthetase Complex to Enhance Viral Replication. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.W.; Huang, Y.P. The RNA degradation pathway is involved in PPARα-modulated anti-oral tumorigenesis. BioMedicine 2019, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Schelker, M.; Mair, C.M.; Jolmes, F.; Welke, R.W.; Klipp, E.; Herrmann, A.; Flöttmann, M.; Sieben, C. Viral RNA Degradation and Diffusion Act as a Bottleneck for the Influenza a Virus Infection Efficiency. PLoS Comput. Biol. 2016, 12, e1005075. [Google Scholar] [CrossRef] [PubMed]
- Panchariya, L.; Khan, W.A.; Kuila, S.; Sonkar, K.; Sahoo, S.; Ghoshal, A.; Kumar, A.; Verma, D.K.; Hasan, A.; Khan, M.A.; et al. Zinc2+ ion inhibits SARS-CoV-2 main protease and viral replication in vitro. Chem. Commun. 2021, 57, 10083–10086. [Google Scholar] [CrossRef] [PubMed]
- MMüller, F.; Svardal, A.M.; Aukrust, P.; Berge, R.K.; Ueland, P.M.; Frøland, S.S. Elevated plasma concentration of reduced homocysteine in patients with human immunodeficiency virus infection. Am. J. Clin. Nutr. 1996, 63, 242–248. [Google Scholar] [CrossRef]
- Korenaga, M.; Nishina, S.; Korenaga, K.; Tomiyama, Y.; Yoshioka, N.; Hara, Y.; Sasaki, Y.; Shimonaka, Y.; Hino, K. Branched-chain amino acids reduce hepatic iron accumulation and oxidative stress in hepatitis C virus polyprotein-expressing mice. Liver Int. Off. J. Int. Assoc. Study Liver 2015, 35, 1303–1314. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Chen, Y.H.; Huang, W.R.; Huang, J.W.; Chen, I.C.; Chang, Y.K.; Wang, C.Y.; Chang, C.D.; Liao, T.L.; Nielsen, B.L.; et al. Oncolytic avian reovirus σA-modulated fatty acid metabolism through the PSMB6/Akt/SREBP1/acetyl-CoA carboxylase pathway to increase energy production for virus replication. Vet. Microbiol. 2022, 273, 109545. [Google Scholar] [CrossRef]
- Toohey, J.I. Sulfur metabolism in AIDS: Cystamine as an anti-HIV agent. AIDS Res. Hum. Retroviruses 2009, 25, 1057–1060. [Google Scholar] [CrossRef]
- Breitkreutz, R.; Holm, S.; Pittack, N.; Beichert, M.; Babylon, A.; Yodoi, J.; Dröge, W. Massive loss of sulfur in HIV infection. AIDS Res. Hum. Retroviruses 2000, 16, 203–209. [Google Scholar] [CrossRef]
- Hinton, A.C.; Hammock, B.D. Juvenile hormone esterase (JHE) from Tenebrio molitor: Full-length cDNA sequence, in vitro expression, and characterization of the recombinant protein. Insect Biochem. Mol. Biol. 2003, 33, 477–487. [Google Scholar] [CrossRef]
- Jones, G.; Venkataraman, V.; Ridley, B.; O’Mahony, P.; Turner, H. Structure, expression and gene sequence of a juvenile hormone esterase-related protein from metamorphosing larvae of Trichoplusia ni. Biochem. J. 1994, 302 Pt 3, 827–835. [Google Scholar] [CrossRef]
- Bai, H.; Ramaseshadri, P.; Palli, S.R. Identification and characterization of juvenile hormone esterase gene from the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 2007, 37, 829–837. [Google Scholar] [CrossRef]
- Campbell, P.M.; Harcourt, R.L.; Crone, E.J.; Claudianos, C.; Hammock, B.D.; Russell, R.J.; Oakeshott, J.G. Identification of a juvenile hormone esterase gene by matching its peptide mass fingerprint with a sequence from the Drosophila genome project. Insect Biochem. Mol. Biol. 2001, 31, 513–520. [Google Scholar] [CrossRef]
- Vermunt, A.M.; Koopmanschap, A.B.; Vlak, J.M.; de Kort, C.A. Cloning and sequence analysis of cDNA encoding a putative juvenile hormone esterase from the Colorado potato beetle. Insect Biochem. Mol. Biol. 1997, 27, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, X.; Shiotsuki, T.; Wang, Z.; Xu, X.; Huang, Y.; Li, M.; Li, K.; Tan, A. Depletion of juvenile hormone esterase extends larval growth in Bombyx mori. Insect Biochem. Mol. Biol. 2017, 81, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Mackert, A.; Hartfelder, K.; Bitondi, M.M.; Simões, Z.L. The juvenile hormone (JH) epoxide hydrolase gene in the honey bee (Apis mellifera) genome encodes a protein which has negligible participation in JH degradation. J. Insect Physiol. 2010, 56, 1139–1146. [Google Scholar] [CrossRef] [PubMed]




| Sample | RawReads | RawBases | CleanReads | ValidBases | Q30 | GC |
|---|---|---|---|---|---|---|
| CK_1 | 48.33 M | 7.25G | 47.27 M | 94.87% | 87.79% | 49.76% |
| CK_2 | 48.26 M | 7.24G | 47.24 M | 95.23% | 90.05% | 49.65% |
| CK_3 | 48.54 M | 7.28G | 47.54 M | 95.64% | 90.36% | 49.31% |
| MbMNPV_1 | 50.64 M | 7.60G | 49.49 M | 95.07% | 89.39% | 49.47% |
| MbMNPV_2 | 49.58 M | 7.44G | 48.39 M | 95.04% | 90.02% | 48.86% |
| MbMNPV_3 | 50.75 M | 7.61G | 49.58 M | 94.99% | 89.25% | 49.82% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Dai, J.; Zhang, G.; Singh, D.; Zhang, X.; Liang, Z. Mamestra brassicae Multiple Nucleopolyhedroviruses Prevents Pupation of Helicoverpa armigera by Regulating Juvenile Hormone Titer. Insects 2024, 15, 202. https://doi.org/10.3390/insects15030202
Yang Y, Dai J, Zhang G, Singh D, Zhang X, Liang Z. Mamestra brassicae Multiple Nucleopolyhedroviruses Prevents Pupation of Helicoverpa armigera by Regulating Juvenile Hormone Titer. Insects. 2024; 15(3):202. https://doi.org/10.3390/insects15030202
Chicago/Turabian StyleYang, Yanqing, Jinping Dai, Guozhi Zhang, Deepali Singh, Xiaoxia Zhang, and Zhenpu Liang. 2024. "Mamestra brassicae Multiple Nucleopolyhedroviruses Prevents Pupation of Helicoverpa armigera by Regulating Juvenile Hormone Titer" Insects 15, no. 3: 202. https://doi.org/10.3390/insects15030202




