Comparing Survival of Israeli Acute Paralysis Virus Infection among Stocks of U.S. Honey Bees
Abstract
Simple Summary
Abstract
1. Introduction
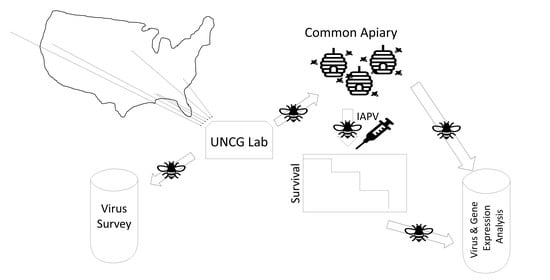
2. Materials and Methods
2.1. Honey Bee Stocks
2.2. Experimental Treatment, Survival Analysis, and Sampling
2.3. RNA Extraction and cDNA Synthesis
2.4. Quantification of Viruses
2.5. Quantification of Immune Genes
3. Results
3.1. Virus Survey of Stocks
3.2. Variation in IAPV-Inoculated Worker Survival among Honey Bee Colonies and Stocks
3.3. Correlation of IAPV-Inoculated Worker Survival with Immune Gene Expression
3.4. Relation of IAPV-Inoculated Worker Survival to Virus Titers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hung, K.-L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B 2018, 285, 20172140. [Google Scholar] [CrossRef] [PubMed]
- Mullin, C.A.; Frazier, M.; Frazier, J.L.; Ashcraft, S.; Simonds, R.; vanEngelsdorp, D.; Pettis, J.S. High levels of miticides and agrochemicals in North American apiaries: Implications for honey bee health. PLoS ONE 2010, 5, e9754. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.M. Honey bee toxicology. Annu. Rev. Entomol. 2015, 60, 415–434. [Google Scholar] [CrossRef] [PubMed]
- Goulson, D.; Nicholls, E.; Botias, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef]
- Simone-Finstrom, M.; Li-Byarlay, H.; Huang, M.H.; Strand, M.K.; Rueppell, O.; Tarpy, D.R. Migratory management and environmental conditions affect lifespan and oxidative stress in honey bees. Sci. Rep. 2016, 6, 32023. [Google Scholar] [CrossRef]
- Krupke, C.H.; Hunt, G.J.; Eitzer, B.D.; Andino, G.; Given, K. Multiple routes of pesticide exposure for honey bees living near agricultural fields. PLoS ONE 2012, 7, e29268. [Google Scholar] [CrossRef]
- DeGrandi-Hoffman, G.; Chen, Y. Nutrition, immunity and viral infections in honey bees. Curr. Opin. Insect Sci. 2015, 10, 170–176. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Flenniken, M.L. Bee viruses: Ecology, pathogenicity, and impacts. Annu. Rev. Entomol. 2019, 64, 205–226. [Google Scholar] [CrossRef]
- VanEngelsdorp, D.; Speybroeck, N.; Evans, J.D.; Kim Nguyen, B.; Mullin, C.; Frazier, M.; Frazier, J.; Cox-Foster, D.; Chen, Y.; Tarpy, D.R.; et al. Weighing risk factors associated with bee colony collapse disorder by classification and regression tree analysis. J. Econ. Entomol. 2010, 103, 1517–1523. [Google Scholar] [CrossRef]
- Di Prisco, G.; Annoscia, D.; Margiotta, M.; Ferrara, R.; Varricchio, P.; Zanni, V.; Caprio, E.; Nazzi, F.; Pennacchio, F. A mutualistic symbiosis between a parasitic mite and a pathogenic virus undermines honey bee immunity and health. Proc. Natl. Acad. Sci. USA 2016, 113, 3203–3208. [Google Scholar] [CrossRef]
- Wilfert, L.; Long, G.; Leggett, H.; Schmid-Hempel, P.; Butlin, R.; Martin, S.; Boots, M. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 2016, 351, 594–597. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.P.; Siede, R. Honey bee viruses. In Advances in Virus Research; Maramorosch, K., Shatkin, A.J., Murphy, F.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; Volume 70, pp. 33–80. [Google Scholar]
- McMenamin, A.J.; Genersch, E. Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 2015, 8, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Brutscher, L.M.; McMenamin, A.J.; Flenniken, M.L. The Buzz about Honey Bee Viruses. PLoS Pathog. 2016, 12, e1005757. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, A.J.; Flenniken, M.L. Recently identified bee viruses and their impact on bee pollinators. Curr. Opin. Insect Sci. 2018, 26, 120–129. [Google Scholar] [CrossRef]
- Amiri, E.; Strand, M.K.; Tarpy, D.R.; Rueppell, O. Honey bee queens and virus infections. Viruses 2020, 12, 322. [Google Scholar] [CrossRef]
- Bailey, L.; Ball, B.V. Honey Bee Pathology; Academic Press: London, UK, 1991. [Google Scholar]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Phenix-Lan, Q.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef]
- Chen, Y.P.; Pettis, J.S.; Corona, M.; Chen, W.P.; Li, C.J.; Spivak, M.; Visscher, P.K.; DeGrandi-Hoffman, G.; Boncristiani, H.; Zhao, Y.; et al. Israeli acute paralysis virus: Epidemiology, pathogenesis and implications for honey bee health. PLoS Pathog. 2014, 10, e1004261. [Google Scholar] [CrossRef]
- Boncristiani, H.F.; Evans, J.D.; Chen, Y.; Pettis, J.; Murphy, C.; Lopez, D.L.; Simone-Finstrom, M.; Strand, M.; Tarpy, D.R.; Rueppell, O. In-vitro infection of pupae with Israeli Acute Paralysis Virus suggests variation for susceptibility and disturbance of transcriptional homeostasis in honey bees (Apis mellifera). PLoS ONE 2013, 8, e73429. [Google Scholar] [CrossRef]
- Amiri, E.; Seddon, G.; Zuluaga Smith, W.; Strand, M.K.; Tarpy, D.R.; Rueppell, O. Israeli acute paralysis virus: Honey bee queen–worker interaction and potential virus transmission pathways. Insects 2019, 10, 9. [Google Scholar] [CrossRef]
- Geffre, A.C.; Gernat, T.; Harwood, G.P.; Jones, B.M.; Gysi, D.M.; Hamilton, A.R.; Bonning, B.C.; Toth, A.L.; Robinson, G.E.; Dolezal, A.G. Honey bee virus causes context-dependent changes in host social behavior. Proc. Natl. Acad. Sci. USA 2020, 117, 10406–10413. [Google Scholar] [CrossRef]
- Maori, E.; Paldi, N.; Shafir, S.; Kalev, H.; Tsur, E.; Glick, E.; Sela, I. IAPV, a bee-affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Mol. Biol. 2009, 18, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.P.; Powell, J.E.; Perutka, J.; Geng, P.; Heckmann, L.C.; Horak, R.D.; Davies, B.W.; Ellington, A.D.; Barrick, J.E.; Moran, N.A. Engineered symbionts activate honey bee immunity and limit pathogens. Science 2020, 367, 573–576. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, D.C.; Laget, D.; De Smet, L.; Claeys Boúúaert, D.; Brunain, M.; Veerkamp, R.F.; Brascamp, E.W. Heritability estimates of the novel trait ‘suppressed in ovo virus infection’ in honey bees (Apis mellifera). Sci. Rep. 2020, 10, 14310. [Google Scholar] [CrossRef] [PubMed]
- Wilson-Rich, N.; Spivak, M.; Fefferman, N.H.; Starks, P.T. Genetic, individual, and group facilitation of disease resistance in insect societies. Annu. Rev. Entomol. 2009, 54, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Cremer, S.; Armitage, S.A.O.; Schmid-Hempel, P. Social immunity. Curr. Biol. 2007, 17, R693–R702. [Google Scholar] [CrossRef] [PubMed]
- Maori, E.; Garbian, Y.; Kunik, V.; Mozes-Koch, R.; Malka, O.; Kalev, H.; Sabath, N.; Sela, I.; Shafir, S. A transmissible RNA pathway in honey bees. Cell Rep. 2019, 27, 1949–1959.e6. [Google Scholar] [CrossRef]
- Shorter, J.R.; Rueppell, O. A review on self-destructive defense behaviors in social insects. Ins. Soc. 2012, 59, 1–10. [Google Scholar] [CrossRef]
- Spivak, M.; Reuter, G.S. Performance of hygienic honey bee colonies in a commercial apiary. Apidologie 1998, 29, 291–302. [Google Scholar] [CrossRef]
- Arechavaleta-Velasco, M.E.; Alcala-Escamilla, K.; Robles-Rios, C.; Tsuruda, J.M.; Hunt, G.J. Fine-scale linkage mapping reveals a small set of candidate genes influencing honey bee grooming behavior in response to Varroa mites. PLoS ONE 2012, 7, e47269. [Google Scholar] [CrossRef]
- Simone, M.; Evans, J.D.; Spivak, M. Resin collection and social immunity in honey bees. Evolution 2009, 63, 3016–3022. [Google Scholar] [CrossRef]
- Simone-Finstrom, M. Social immunity and the superorganism: Behavioral defenses protecting honey bee colonies from pathogens and parasites. Bee World 2017, 94, 21–29. [Google Scholar] [CrossRef]
- Cremer, S.; Pull, C.D.; Fürst, M.A. Social immunity: Emergence and evolution of colony-level disease protection. Annu. Rev. Entomol. 2018, 63, 105–123. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.-L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Antiviral defense mechanisms in honey bees. Curr. Opin. Insect Sci. 2015, 10, 71–82. [Google Scholar] [CrossRef]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Virus and dsRNA-triggered transcriptional responses reveal key components of honey bee antiviral defense. Sci. Rep. 2017, 7, 6448. [Google Scholar] [CrossRef]
- Brutscher, L.M.; Flenniken, M.L. RNAi and antiviral defense in the honey bee. J. Immunol. Res. 2015, 2015, 941897. [Google Scholar] [CrossRef]
- Meixner, M.D.; Pinto, M.A.; Bouga, M.; Kryger, P.; Ivanova, E.; Fuchs, S. Standard methods for characterising subspecies and ecotypes of Apis mellifera. J. Apicult. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- Cobey, S.; Sheppard, W.S.; Tarpy, D.R. Status of breeding practices and genetic diversity in domestic US honey bees. In Honey Bee Colony Health; Sammataro, D., Yoder, J.A., Eds.; Challenges and Sustainable Solutions; CRC: Boca Raton, FL, USA, 2012; pp. 39–49. [Google Scholar]
- Saelao, P.; Simone-Finstrom, M.; Avalos, A.; Bilodeau, L.; Danka, R.; de Guzman, L.; Rinkevich, F.; Tokarz, P. Genome-wide patterns of differentiation within and among U.S. commercial honey bee stocks. BMC Genom. 2020, 21, 704. [Google Scholar] [CrossRef]
- Tarpy, D.R. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc. R. Soc. Lond. B 2003, 270, 99–103. [Google Scholar] [CrossRef]
- Oddie, M.; Büchler, R.; Dahle, B.; Kovacic, M.; Le Conte, Y.; Locke, B.; De Miranda, J.R.; Mondet, F.; Neumann, P. Rapid parallel evolution overcomes global honey bee parasite. Sci. Rep. 2018, 8, 7704. [Google Scholar] [CrossRef]
- Le Conte, Y.; Meixner, M.D.; Brandt, A.; Carreck, N.L.; Costa, C.; Mondet, F.; Büchler, R. Geographical Distribution and selection of European honey bees resistant to Varroa destructor. Insects 2020, 11, 873. [Google Scholar] [CrossRef] [PubMed]
- Locke, B.; Forsgren, E.; de Miranda, J. Increased tolerance and resistance to virus infections: A possible factor in the survival of Varroa destructor. PLoS ONE 2014, 9, e99998. [Google Scholar] [CrossRef] [PubMed]
- Thaduri, S.; Stephan, J.G.; de Miranda, J.R.; Locke, B. Disentangling host-parasite-pathogen interactions in a varroa-resistant honeybee population reveals virus tolerance as an independent, naturally adapted survival mechanism. Sci. Rep. 2019, 9, 6221. [Google Scholar] [CrossRef] [PubMed]
- Rothenbuhler, W.C. Behaviour genetics of nest cleaning in honey bees. IV. Responses of F1 and backcross generations to disease-killed brood. Am. Zool. 1964, 4, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Boecking, O.; Bienefeld, K.; Drescher, W. Heritability of the Varroa-specific hygienic behaviour in honey bees (Hymenoptera: Apidae). J. Anim. Breed. Genet. 2000, 117, 417–424. [Google Scholar] [CrossRef]
- Mondet, F.; Beaurepaire, A.; McAfee, A.; Locke, B.; Alaux, C.; Blanchard, S.; Danka, B.; Le Conte, Y. Honey bee survival mechanisms against the parasite Varroa destructor: A systematic review of phenotypic and genomic research efforts. Int. J. Parasitol. 2020, 50, 433–447. [Google Scholar] [CrossRef]
- Tsuruda, J.M.; Harris, J.W.; Bourgeois, L.; Danka, R.G.; Hunt, G.J. High-resolution linkage analyses to identify genes that influence Varroa Sensitive Hygiene behavior in honey bees. PLoS ONE 2012, 7, e48276. [Google Scholar] [CrossRef]
- Behrens, D.; Huang, Q.; Gessner, C.; Rosenkranz, P.; Frey, E.; Locke, B.; Moritz, R.F.; Kraus, F.B. Three QTL in the honey bee Apis mellifera L. suppress reproduction of the parasitic mite Varroa destructor. Ecol. Evol. 2011, 1, 451–458. [Google Scholar] [CrossRef]
- Spivak, M.; Reuter, G.S.; Lee, K.; Ranum, B. The future of the MN Hygienic Stock of bees is in good hands! Am. Bee J. 2009, 149, 965–967. [Google Scholar]
- Danka, R.G.; Harris, J.W.; Dodds, G.E. Selection of VSH-derived “Pol-line” honey bees and evaluation of their Varroa-resistance characteristics. Apidologie 2016, 47, 483–490. [Google Scholar] [CrossRef]
- Harris, J.; Rinderer, T. Varroa resistance of hybrid ARS Russian honey bees. Am. Bee J. 2004, 144, 797–800. [Google Scholar]
- Kulincevic, J.M.; Rothenbuhler, W.C. Selection for resistance and susceptibility to hairless-black syndrome in the honeybee. J. Invertebr. Pathol. 1975, 25, 289–295. [Google Scholar] [CrossRef]
- Decanini, L.I.; Collins, A.M.; Evans, J.D. Variation and heritability in immune gene expression by diseased honeybees. J. Hered. 2007, 98, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Khongphinitbunjong, K.; de Guzman, L.I.; Tarver, M.R.; Rinderer, T.E.; Chen, Y.; Chantawannakul, P. Differential viral levels and immune gene expression in three stocks of Apis mellifera induced by different numbers of Varroa destructor. J. Insect Physiol. 2015, 72, 28–34. [Google Scholar] [CrossRef]
- Kevill, J.; Lee, K.; Goblirsch, M.; McDermott, E.; Tarpy, D.R.; Spivak, M.; Schroeder, D.C. The pathogen profile of a honey bee queen does not reflect that of her workers. Insects 2020, 11, 382. [Google Scholar] [CrossRef]
- Yue, C.; Schroder, M.; Gisder, S.; Genersch, E. Vertical-transmission routes for deformed wing virus of honeybees (Apis mellifera). J. Gen. Virol. 2007, 88, 2329–2336. [Google Scholar] [CrossRef]
- Evans, J.D.; Chen, Y.P.; Prisco, G.D.; Pettis, J.; Williams, V. Bee cups: Single-use cages for honey bee experiments. J. Apic. Res. 2009, 48, 300–302. [Google Scholar] [CrossRef]
- Kuster, R.D.; Boncristiani, H.F.; Rueppell, O. Immunogene and viral transcript dynamics during parasitic Varroa destructor mite infection of developing honey bee (Apis mellifera) pupae. J. Exp. Biol. 2014, 217, 1710–1718. [Google Scholar] [CrossRef]
- Amiri, E.; Kryger, P.; Meixner, M.; Strand, M.K.; Tarpy, D.R.; Rueppell, O. Quantitative patterns of vertical transmission of deformed wing virus in honey bees. PLoS ONE 2018, 13, e0195283. [Google Scholar] [CrossRef]
- Di Prisco, G.; Pennacchio, F.; Caprio, E.; Boncristiani, H.F.; Evans, J.D.; Chen, Y.P. Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 2011, 92, 151–155. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Amiri, E.; Meixner, M.D.; Kryger, P. Deformed wing virus can be transmitted during natural mating in honey bees and infect the queens. Sci. Rep. 2016, 6, 33065. [Google Scholar] [CrossRef] [PubMed]
- Traynor, K.S.; Rennich, K.; Forsgren, E.; Rose, R.; Pettis, J.; Kunkel, G.; Madella, S.; Evans, J.; Lopez, D.; vanEngelsdorp, D. Multiyear survey targeting disease incidence in US honey bees. Apidologie 2016, 47, 325–347. [Google Scholar] [CrossRef]
- Ravoet, J.; De Smet, L.; Wenseleers, T.; de Graaf, D.C. Vertical transmission of honey bee viruses in a Belgian queen breeding program. BMC Vet. Res. 2015, 11, 61. [Google Scholar] [CrossRef]
- Chen, Y.P.; Evans, J.; Feldlaufer, M. Horizontal and vertical transmission of viruses in the honeybee, Apis mellifera. J. Invertebr. Pathol. 2006, 92, 152–159. [Google Scholar] [CrossRef]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Patterns of viral infection in honey bee queens. J. Gen. Virol. 2013, 94, 668–676. [Google Scholar] [CrossRef]
- Neumann, P.; Blacquière, T. The Darwin cure for apiculture? Natural selection and managed honeybee health. Evol. Appl. 2017, 10, 226–230. [Google Scholar] [CrossRef]
- Al Naggar, Y.; Paxton, R.J. Mode of transmission determines the virulence of Black queen cell virus in adult honey bees, posing a future threat to bees and apiculture. Viruses 2020, 12, 535. [Google Scholar] [CrossRef]
- Bull, J.C.; Ryabov, E.V.; Prince, G.; Mead, A.; Zhang, C.; Baxter, L.A.; Pell, J.K.; Osborne, J.L.; Chandler, D. A strong immune response in young adult honeybees masks their increased susceptibility to infection compared to older bees. PLoS Pathog. 2012, 8, e1003083. [Google Scholar] [CrossRef]
- Galbraith, D.A.; Yang, X.; Niño, E.L.; Yi, S.; Grozinger, C.; Schneider, D.S. Parallel epigenomic and transcriptomic responses to viral infection in honey bees (Apis mellifera). PLoS Pathog. 2015, 11, e1004713. [Google Scholar] [CrossRef]
- Wang, H.; Meeus, I.; Smagghe, G. Israeli acute paralysis virus associated paralysis symptoms, viral tissue distribution and Dicer-2 induction in bumblebee workers (Bombus terrestris). J. Gen. Virol. 2016, 97, 1981–1989. [Google Scholar] [CrossRef] [PubMed][Green Version]



| CAH | MHY | POL | RUS | |
|---|---|---|---|---|
| ITC | 0.91 ± 0.15 p = 0.518 | 1.14 ± 0.31 p = 0.681 | 0.87 ± 0.17 p = 0.416 | 0.64 ± 0.14 p = 0.002 |
| CAH | 1.25 ± 0.30 p = 0.459 | 0.61 ± 0.17 p = 0.004 | 0.71 ± 0.12 p = 0.004 | |
| MHY | 0.49 ± 0.32 p = 0.026 | 0.56 ± 0.30 p = 0.057 | ||
| POL | 1.62 ± 0.20 p = 0.015 |
| ITC | CAH | MHY | POL | |
|---|---|---|---|---|
| RUS | 1.77 ± 0.14 p < 0.001 | 0.99 ± 0.12 p = 0.945 | 2.29 ± 0.30 p = 0.006 | 0.61 ± 0.17 p = 0.004 |
| POL | 1.40 ± 0.22 p = 0.119 | 0.78 ± 0.20 p = 0.233 | 1.81 ± 0.34 p = 0.081 | |
| MHY | 0.77 ± 0.31 p = 0.409 | 0.43 ± 0.30 p = 0.006 | ||
| CAH | 1.79 ± 0.15 p < 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhatia, S.; Baral, S.S.; Vega Melendez, C.; Amiri, E.; Rueppell, O. Comparing Survival of Israeli Acute Paralysis Virus Infection among Stocks of U.S. Honey Bees. Insects 2021, 12, 60. https://doi.org/10.3390/insects12010060
Bhatia S, Baral SS, Vega Melendez C, Amiri E, Rueppell O. Comparing Survival of Israeli Acute Paralysis Virus Infection among Stocks of U.S. Honey Bees. Insects. 2021; 12(1):60. https://doi.org/10.3390/insects12010060
Chicago/Turabian StyleBhatia, Shilpi, Saman S. Baral, Carlos Vega Melendez, Esmaeil Amiri, and Olav Rueppell. 2021. "Comparing Survival of Israeli Acute Paralysis Virus Infection among Stocks of U.S. Honey Bees" Insects 12, no. 1: 60. https://doi.org/10.3390/insects12010060
APA StyleBhatia, S., Baral, S. S., Vega Melendez, C., Amiri, E., & Rueppell, O. (2021). Comparing Survival of Israeli Acute Paralysis Virus Infection among Stocks of U.S. Honey Bees. Insects, 12(1), 60. https://doi.org/10.3390/insects12010060