Analysis of miRNAs in the Heads of Different Castes of the Bumblebee Bombus lantschouensis (Hymenoptera: Apidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. RNA Isolation, Library Construction, and Sequencing
2.3. Read Filtering and Mapping
2.4. Prediction of miRNAs
2.5. Differential Expression Analysis of miRNAs
2.6. Target Gene Prediction and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analyses
2.7. miRNA Extraction and RT-qPCR
3. Results
3.1. Analysis of Sequencing Data
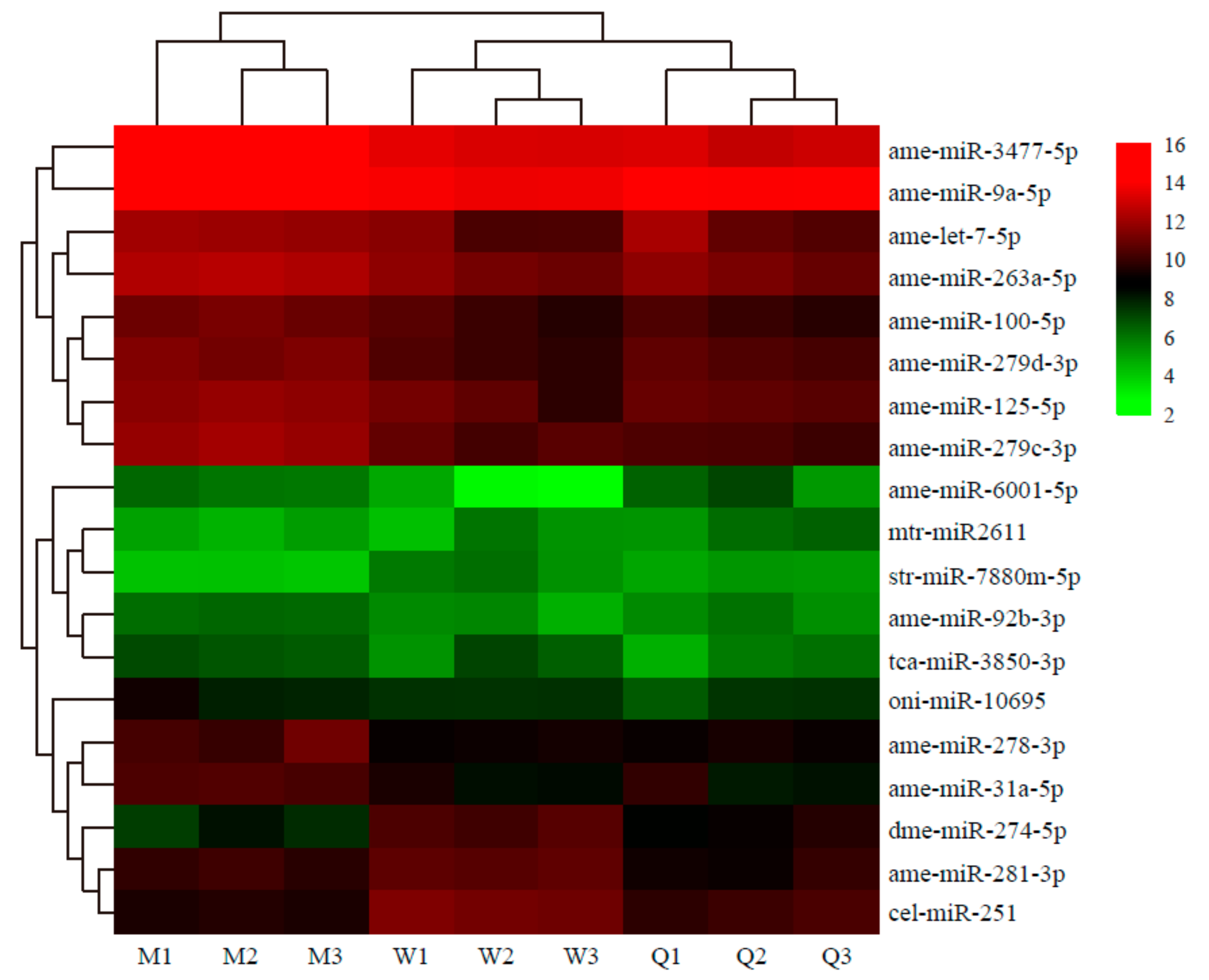
3.2. Identification and Expression of Known and Novel miRNAs
3.3. DE miRNAs in Males, Queens, and Workers
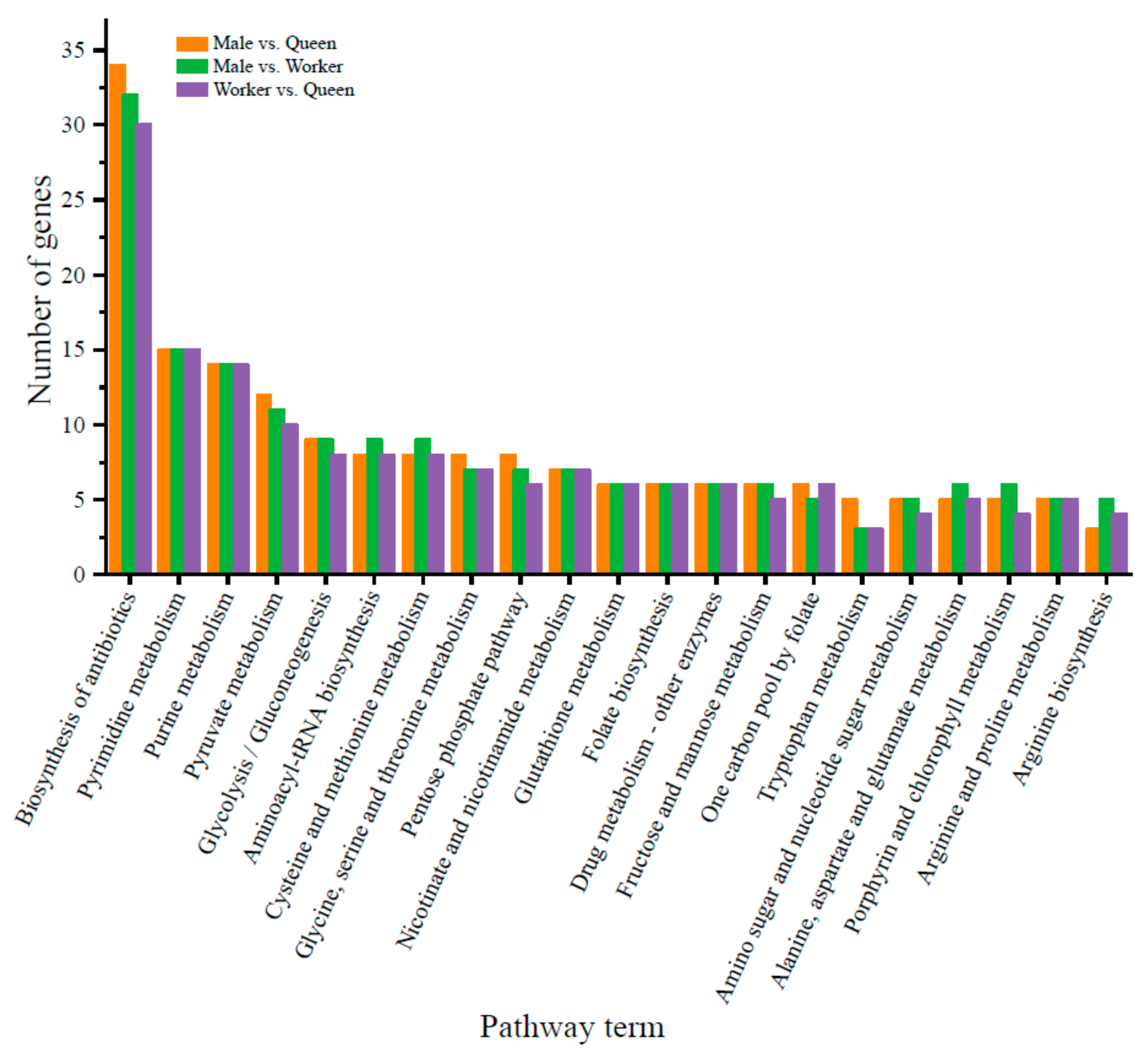
3.4. Prediction of Target Genes of DE miRNAs and Functional Enrichment Analysis
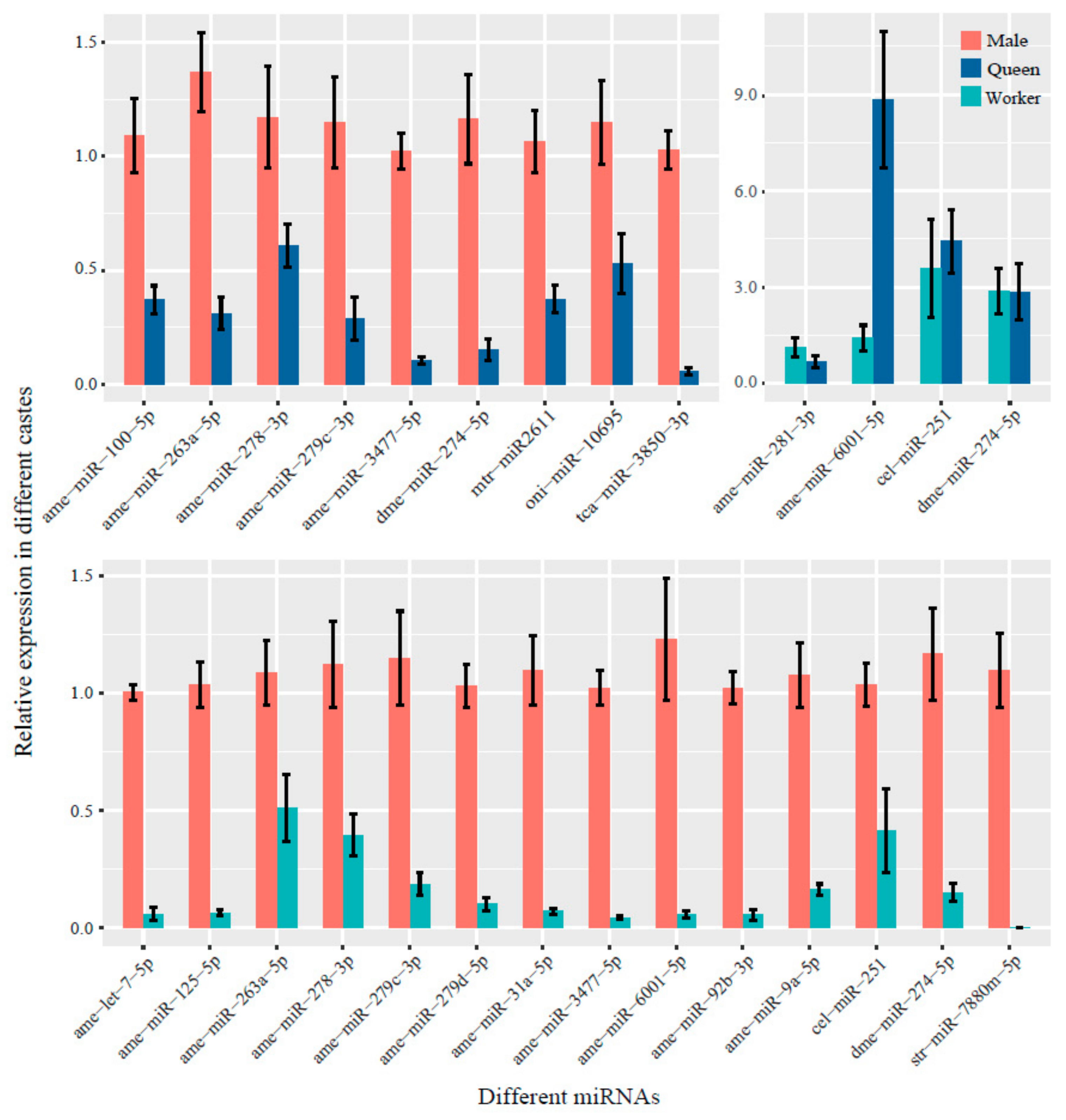
3.5. Validation of Differential miRNA Expression by RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- An, J.; Huang, J.; Shao, Y.; Zhang, S.; Wang, B.; Liu, X.; Wu, J.; Williams, P.H. The bumblebees of North China (Apidae, Bombus Latreille). Zootaxa 2014, 3830, 1–89. [Google Scholar] [CrossRef]
- Huang, J.; An, J. Species diversity, pollination application and strategy for conservation of the bumblebees of China. Biodivers. Sci. 2018, 26, 486–497. [Google Scholar] [CrossRef]
- Velthuis, H.H.W.; van Doorn, A. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie 2006, 37, 421–451. [Google Scholar] [CrossRef] [Green Version]
- Goulson, D.; Lye, G.C.; Darvill, B. Decline and conservation of bumble bees. Annu. Rev. Entomol. 2008, 53, 191–208. [Google Scholar] [CrossRef]
- Thorp, R.W.; Horning, D.S.; Dunning, L.L. Bumble bees and cuckoo bumble bees of California (Hyenoptera, Apidae). Bull. Calif. Insect Surv. 1983, 23, 1–79. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucas, K.; Raikhel, A.S. Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem. Mol. 2013, 43, 24–38. [Google Scholar] [CrossRef]
- Behura, S.K. Insect microRNAs: Structure, function and evolution. Insect Biochem. Mol. 2007, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Menzel, R. Searching for the memory trace in a mini-brain, the honeybee. Learn. Mem. 2001, 8, 53–62. [Google Scholar] [CrossRef]
- Whitfield, C.W.; Ben-Shahar, Y.; Brillet, C.; Leoncini, I.; Crauser, D.; Leconte, Y.; Rodriguez-Zas, S.; Robinson, G.E. Genomic Dissection of Behavioral Maturation in the Honey Bee. Proc. Natl. Acad. Sci. USA 2006, 103, 16068–16075. [Google Scholar] [CrossRef] [PubMed]
- Ashby, R.; Forêt, S.; Searle, I.; Maleszka, R. MicroRNAs in Honey Bee Caste Determination. Sci. Rep. UK 2016, 6, 18794. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, F.; Li, W.; Li, Z.; Pan, J.; Yan, L.; Zhang, S.; Huang, Z.Y.; Su, S. Differences in microRNAs and their expressions between foraging and dancing honey bees, Apis mellifera L. J. Insect Physiol. 2012, 58, 1438–1443. [Google Scholar] [CrossRef] [PubMed]
- Macedo, L.M.; Nunes, F.M.; Freitas, F.C.; Pires, C.V.; Tanaka, E.D.; Martins, J.R.; Piulachs, M.D.; Cristino, A.S.; Pinheiro, D.G.; Simoes, Z.L. MicroRNA signatures characterizing caste-independent ovarian activity in queen and worker honeybees (Apis mellifera L.). Insect Mol. Biol. 2016, 25, 216–226. [Google Scholar] [CrossRef]
- Sun, D.T.; Dong, J.; Huang, J.X.; He, S.Y.; Wu, J. Solexa sequencing and bioinformatics analysis of microRNA in the queen head of the bumblebee Bombus lantschouensis. Acta Entomol. Sin. 2015, 58, 1291–1299. [Google Scholar]
- Dong, J.; Li, J.; Huang, J.; Wu, J. Identification of suitable reference genes for miRNA quantitation in bumblebee (Hymenoptera: Apidae) response to reproduction. Apidologie 2019, 50, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Collins, D.H.; Mohorianu, I.; Beckers, M.; Moulton, V.; Dalmay, T.; Bourke, A.F.G. MicroRNAs Associated with Caste Determination and Differentiation in a Primitively Eusocial Insect. Sci. Rep. UK 2017, 7, 45674. [Google Scholar] [CrossRef] [PubMed]
- Goulson, D.; Hanley, M.E. Distribution and forage use of exotic bumblebees in South Island, New Zealand. N. Z. J. Ecol. 2004, 28, 225–232. [Google Scholar]
- Schmid-Hempel, P.; Schmid-Hempel, R.; Brunner, P.C.; Seeman, O.D.; Allen, G.R. Invasion success of the bumblebee, Bombus terrestris, despite a drastic genetic bottleneck. Heredity 2007, 99, 414–422. [Google Scholar] [CrossRef]
- Inari, N.; Nagamitsu, T.; Kenta, T.; Goka, K.; Hiura, T. Spatial and temporal pattern of introduced Bombus terrestris abundance in Hokkaido, Japan, and its potential impact on native bumblebees. Popul. Ecol. 2005, 47, 77–82. [Google Scholar] [CrossRef]
- Schmid-Hempel, R.; Eckhardt, M.; Goulson, D.; Heinzmann, D.; Lange, C.; Plischuk, S.; Escudero, L.R.; Salathé, R.; Scriven, J.J.; Schmid-Hempel, P. The invasion of southern South America by imported bumblebees and associated parasites. J. Anim. Ecol. 2014, 83, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Yuan, X.; Huang, J.; An, J. Habitat suitability for the invasion of Bombus terrestris in East Asian countries: A case study of spatial overlap with local Chinese bumblebees. Sci. Rep. UK 2018, 8, 11010–11035. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, Z.; Huang, J.; Yuan, X.; Ding, G.; An, J. Queen traits and colony size of four bumblebee species of China. Insectes Soc. 2018, 65, 537–547. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, Z.; An, J. Pollen Release Dynamics and Daily Patterns of Pollen-Collecting Activity of Honeybee Apis mellifera and Bumblebee Bombus lantschouensis in Solar Greenhouse. Insects 2019, 10, 216. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Cameron, S.A.; Hines, H.M.; Williams, P.H. A comprehensive phylogeny of the bumble bees (Bombus). Biol. J. Linn. Soc. 2007, 91, 161–188. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; et al. Integrated profiling of microRNAs and mRNAs: MicroRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H. TBtools, a Toolkit for Biologists integrating various HTS data handling tools with a user friendly interface. BioRxiv 2018, 289660. [Google Scholar]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol. 2004, 2, 1863–1879. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Gaul, U.; Iovino, N.; Unnerstall, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Mount, S.M.; Gotea, V.; Lin, C.F.; Hernandez, K.; Makalowski, W. Spliceosomal small nuclear RNA genes in 11 insect genomes. RNA 2006, 13, 5–14. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Peng, J.; Hu, J.; Xu, Z.; Xie, W.; Yuan, L. Localized expression pattern of miR-184 in Drosophila. Mol. Biol. Rep. 2011, 38, 355–358. [Google Scholar] [CrossRef]
- Guo, X.; Su, S.; Geir, S.; Li, W.; Li, Z.; Zhang, S.W.; Chen, S.L.; Chen, R.S. Differential expression of miRNAs related to caste differentiation in the honey bee, Apis mellifera. Apidologie 2016, 47, 495–508. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, X.; Li, J.; Liu, P. Identification and comparative profiling of ovarian and testicular microRNAs in the swimming crab Portunus trituberculatus. Gene 2018, 640, 6–13. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Jiang, H.; Ma, J.; Li, J.; Chen, J. Identification and differential expression of microRNAs in testis and ovary of Amur sturgeon (Acipenser schrenckii). Gene 2018, 658, 36–46. [Google Scholar] [CrossRef]
- Huang, J.; Ju, Z.; Li, Q.; Hou, Q.; Wang, C.; Li, J.; Li, R.; Wang, L.; Sun, T.; Hang, S.; et al. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int. J. Biol. Sci. 2011, 7, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Genome-Wide Identification of MicroRNAs and Their Regulation of Transcriptome on Female Caste Determination of Honeybee (Apis Mellifera). Ph.D. Thesis, Zhejiang University, Hangzhou, Zhejiang, China, 2012. [Google Scholar]
- Weng, R.; Cohen, S. Control of Drosophila Type I and Type II central brain neuroblast proliferation by bantam microRNA. Development 2015, 142, 3713–3720. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zheng, H.; Pan, Q.; Wang, Z.; Zeng, Z. Differentially expressed microRNAs between queen and worker larvae of the honey bee (Apis mellifera). Apidologie 2015, 46, 35–45. [Google Scholar] [CrossRef]
- Bizuayehu, T.T.; Babiak, J.; Norberg, B.; Fernandes, J.M.; Johansen, S.D.; Babiak, I. Sex-biased miRNA expression in Atlantic Halibut (Hippoglossus hippoglossus) brain and gonads. Sex. Dev. 2012, 6, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhang, L.; Chen, X. Differential expression analysis of Paralichthys olivaceus microRNAs in adult ovary and testis by deep sequencing. Gen. Comp. Endocrinol. 2014, 204, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Zou, J.; Wang, K.; Su, Y.; Zhu, Y.; Song, C.; Li, G.; Qu, L.; Zhang, H.; Liu, H. High-throughput sequencing reveals Hypothalamic microRNAs as novel partners involved in timing the rapid development of chicken (Gallus gallus) gonads. PLoS ONE 2015, 10, e0129738. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, Y.; Wang, T.; Guan, J.; Luo, Z.; Chen, H.; Wang, X.; Chen, L.; Ma, J.; Mu, Z.; et al. Repertoire of porcine microRNAs in adult ovary and testis by deep sequencing. Int. J. Biol. Sci. 2011, 7, 1045–1055. [Google Scholar] [CrossRef]
- Quah, S.; Breuker, C.J.; Holland, P.W. A diversity of conserved and novel ovarian microRNAs in the speckled wood (Pararge aegeria). PLoS ONE 2015, 10, e0142243. [Google Scholar] [CrossRef]
- Epstein, Y.; Perry, N.; Volin, M.; Zohar-Fux, M.; Braun, R.; Porat-Kuperstein, L.; Toledano, H. miR-9a modulates maintenance and ageing of Drosophila germline stem cells by limiting N-cadherin expression. Nat. Commun. 2017, 8, 600. [Google Scholar] [CrossRef]
- Chawla, G.; Deosthale, P.; Childress, S.; Wu, Y.; Sokol, N.S. A let-7-to-miR-125 microRNA switch regulates neuronal integrity and lifespan in Drosophila. PLoS Genet. 2016, 12, e1006247. [Google Scholar] [CrossRef]
- Jiang, J.; Ge, X.; Li, Z.; Wang, Y.; Song, Q.; Stanley, D.W.; Tan, A.; Huang, Y. MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2013, 43, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, Y.; Yuan, X.; Pan, R.; Yao, W.; Zhong, L.; Song, Q.; Zheng, S.; Wang, Z.; Xu, Q.; et al. Identification of differentially expressed microRNAs during preadipocyte differentiation in Chinese crested duck. Gene 2018, 661, 126–132. [Google Scholar] [CrossRef] [PubMed]



| Name | miRNA Sequence | Primer Sequence |
|---|---|---|
| ame-let-7-5p | UGAGGUAGUAGGUUGUAUAGU | GCGACGCTGAGGTAGTAGGTTGTATAG |
| ame-miR-9a-5p | UCUUUGGUUAUCUAGCUGUAUGA | GACGCTCTTTGGTTATCTAGCTGTATGA |
| ame-miR-31a-5p | AGGCAAGAUGUCGGCAUAGCUGA | AGGCAAGATGTCGGCATAGCTG |
| ame-miR-92b-3p | AAUUGCACCCGUCCCGGCCUGA | AATTGCACCCGTCCCGGC |
| ame-miR-100-5p | AACCCGUAGAUCCGAACUUGUG | GCAACCCGTAGATCCGAACTTGTG |
| ame-miR-125-5p | CCCCUGAGACCCUAACUUGUGA | GCCCCCTGAGACCCTAACTTGTG |
| ame-miR-263a-5p | AAUGGCACUGGAAGAAUUCACG | CAATGGCACTGGAAGAATTCACG |
| ame-miR-278-3p | UCGGUGGGACUUUCGUCCGUUU | TCGGTGGGACTTTCGTCCGT |
| ame-miR-279c-3p | UGACUAGAGUCACACUCGUCCA | GACGCTGACTAGAGTCACACTCGTCC |
| ame-miR-279d-3p | UGACUAGAUCCACACUCAUCCA | CGCTGACTAGATCCACACTCATCCA |
| ame-miR-281-3p | AAGAGAGCUAUCCAUCGACAGU | CGCTGTCATGGAGTTGCTCTCTTTG |
| ame-miR-3477-5p | UAAUCUCAUGCGGUAACUGUGAG | CGCTAATCTCATGCGGTAACTGTGAG |
| ame-miR-6001-5p | UUCUCUUUGGUUGUUACCACU | GACGCTTCTCTTTGGTTGTTACCACT |
| cel-miR-251 | UUAAGUAGUAGUGCCGUAGAUGA | CGACGCTTAAGTAGTAGTGCCGTAGATG |
| dme-miR-274-5p | CUUGUGACCGUAACAACGGGCG | TTGTGACCGTAACAACGGGCG |
| mtr-miR2611 | UAUUUGUCGAGAGUCAUUCUGA | GACGCTATTTGTCGAGAGTCATTCTGA |
| oni-miR-10695 | UAUGUGAUCGCGGAUUUUGUC | GCTATGTGATTGCGGATTTTGTCA |
| str-miR-7880m-5p | UGUCGGUAGCAAAGAGGUGGUAG | GCTGTCGGTAGCAAAGAGGTGGTAG |
| tca-miR-3850-3p | UUCGAGACUACACGCUGAUUUU | GACGCTTCGAGACTACACGCTGATT |
| U6 | GGCCAAGGATGACACGCAAA |
| Male | Queen | Worker | |
|---|---|---|---|
| Adapter | CTGTAGGCACCATCAAT | CTGTAGGCACCATCAAT | CTGTAGGCACCATCAAT |
| Raw reads | 14,326,369 | 14,736,596 | 16,317,406 |
| Clean reads | 13,569,008 | 13,949,908 | 15,413,820 |
| Mapped (%) | 9,717,155 (71.61%) | 9,731,221 (69.76%) | 10,841,052 (70.33%) |
| Unmapped (%) | 3,851,853 (28.39%) | 4,218,687 (30.24%) | 4,572,761 (29.67%) |
| Male | Queen | Worker | ||||
|---|---|---|---|---|---|---|
| Rank | miRNA | Mean Number of Reads | miRNA | Mean Number of Reads | miRNA | Mean Number of Reads |
| 1 | ame-miR-1-3p | 642,846 | ame-miR-1-3p | 989,562 | ame-miR-1-3p | 963,375 |
| 2 | ame-miR-276-3p | 627,171 | ame-miR-276-3p | 650,030 | ame-miR-317-3p | 709,018 |
| 3 | ame-miR-8-3p | 573,082 | ame-miR-317-3p | 638,023 | ame-miR-276-3p | 676,149 |
| 4 | ame-bantam-3p | 525,099 | ame-miR-277-3p | 611,509 | ame-miR-2796-3p | 642,316 |
| 5 | ame-miR-2796-3p | 414,425 | ame-miR-2796-3p | 570,760 | ame-miR-277-3p | 573,485 |
| 6 | ame-miR-317-3p | 396,338 | ame-bantam-3p | 474,566 | ame-bantam-3p | 510,915 |
| 7 | ame-miR-7-5p | 372,830 | ame-miR-8-3p | 355,134 | ame-miR-7-5p | 358,513 |
| 8 | ame-miR-14-3p | 335,963 | ame-miR-7-5p | 270,861 | ame-miR-8-3p | 328,866 |
| 9 | ame-miR-277-3p | 323,674 | ame-miR-184-3p | 262,970 | ame-miR-14-3p | 280,422 |
| 10 | ame-miR-9a-5p | 284,686 | ame-miR-34-5p | 203,858 | ame-miR-34-5p | 277,987 |
| Male vs. Queen | Male vs. Worker | Worker vs. Queen | |||||||
|---|---|---|---|---|---|---|---|---|---|
| DEM | Log2FC | PADJ | Regulated | Log2FC | PADJ | Regulated | Log2FC | PADJ | Regulated |
| ame-miR-100-5p | 1.03 | 0.004484567 | Up | - | - | - | - | - | - |
| ame-miR-263a-5p | 1.10 | 0.003483259 | Up | 1.14 | 0.000195153 | Up | - | - | - |
| ame-miR-278-3p | 1.32 | 0.000692894 | Up | 1.35 | 0.000435914 | Up | - | - | - |
| ame-miR-279c-3p | 1.69 | 8.42568 × 10−15 | Up | 1.41 | 4.02648 × 10−08 | Up | - | - | - |
| ame-miR-3477-5p | 1.76 | 3.03154 × 10−11 | Up | 1.49 | 3.66407 × 10−19 | Up | - | - | - |
| dme-miR-274-5p | −1.33 | 0.003999496 | Down | −2.54 | 3.08025 × 10−15 | Down | 1.21 | 0.003047672 | Up |
| mtr-miR2611 | −1.15 | 0.020339838 | Down | - | - | - | - | - | - |
| oni-miR-10695 | 1.31 | 0.039998779 | Up | - | - | - | - | - | - |
| tca-miR-3850-3p | 1.19 | 0.026213313 | Up | - | - | - | - | - | - |
| ame-let-7-5p | - | - | - | 1.08 | 0.040255941 | Up | - | - | - |
| ame-miR-125-5p | - | - | - | 1.05 | 0.035218532 | Up | - | - | - |
| ame-miR-279d-3p | - | - | - | 1.23 | 2.63402 × 10−05 | Up | - | - | - |
| ame-miR-31a-5p | - | - | - | 1.60 | 0.000681818 | Up | - | - | - |
| ame-miR-6001-5p | - | - | - | 2.37 | 0.013708048 | Up | −2.67 | 0.026701585 | Down |
| ame-miR-92b-3p | - | - | - | 1.01 | 0.027956543 | Up | - | - | - |
| ame-miR-9a-5p | - | - | - | 1.11 | 1.15411 × 10−14 | Up | - | - | - |
| cel-miR-251 | - | - | - | −1.64 | 2.24266 × 10−19 | Down | 1.12 | 2.16263 × 10−06 | Up |
| str-miR-7880m-5p | - | - | - | −1.70 | 3.24877 × 10−06 | Down | - | - | - |
| ame-miR-281-3p | - | - | - | - | - | - | 1.18 | 8.3985 × 10−05 | Up |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Huang, J.; Zhang, G.; Liu, X.; An, J. Analysis of miRNAs in the Heads of Different Castes of the Bumblebee Bombus lantschouensis (Hymenoptera: Apidae). Insects 2019, 10, 349. https://doi.org/10.3390/insects10100349
Liu M, Huang J, Zhang G, Liu X, An J. Analysis of miRNAs in the Heads of Different Castes of the Bumblebee Bombus lantschouensis (Hymenoptera: Apidae). Insects. 2019; 10(10):349. https://doi.org/10.3390/insects10100349
Chicago/Turabian StyleLiu, Meijuan, Jiaxing Huang, Guangshuo Zhang, Xiaofeng Liu, and Jiandong An. 2019. "Analysis of miRNAs in the Heads of Different Castes of the Bumblebee Bombus lantschouensis (Hymenoptera: Apidae)" Insects 10, no. 10: 349. https://doi.org/10.3390/insects10100349





