A Method for Biomarker Directed Survival Prediction in Advanced Non-Small-Cell Lung Cancer Patients Treated with Carboplatin-Based Therapy
Abstract
:1. Introduction
2. Methods
2.1. Patients and Phase III Trials
2.2. Protein Expression Analysis
2.3. Statistical Methods
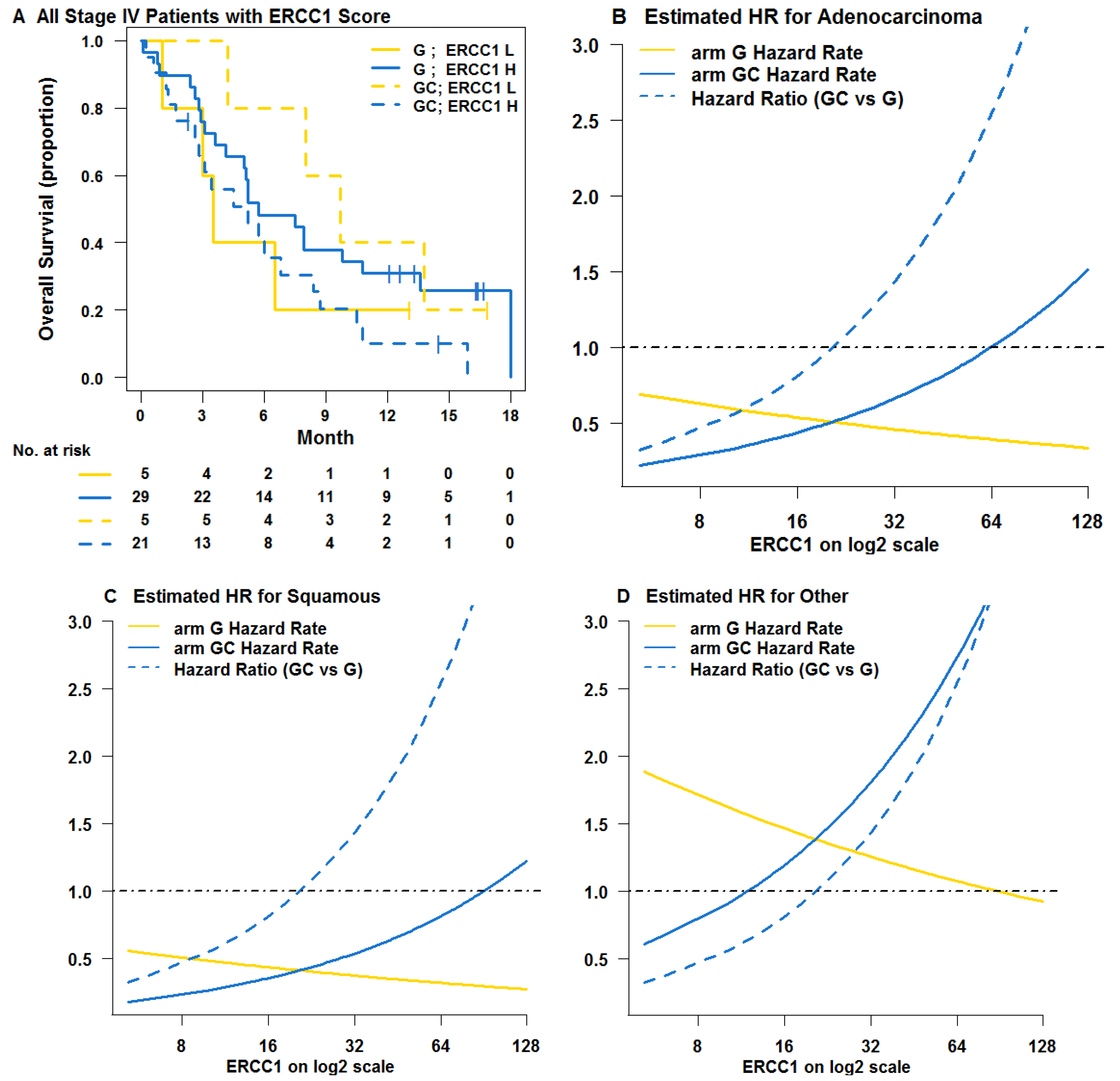
3. Results
3.1. Learning Trial NCT00190710
| Variables | Learning Trial | p | Validation Trial | p | ||
|---|---|---|---|---|---|---|
| G (n = 35) No (%) | GC (n = 34) No (%) | GC (n = 92) No (%) | GC, DC, GD, or DV (n = 183) No (%) | |||
| Age (years) | ||||||
| Median | 75.6 | 70.8 | 0.414 * | 63.2 | 64.3 | 0.204 * |
| Range | 53.1–83.8 | 49.4–85.5 | 39.6–82.6 | 42.1–85.0 | ||
| Sex | ||||||
| Male | 19 (54.3) | 16 (47.1) | 0.633 † | 43 (46.7) | 90 (49.2) | 0.798 † |
| Female | 16 (45.7) | 18 (52.9) | 49 (53.3) | 93 (50.8) | ||
| Histology | ||||||
| Adenocarcinoma | 19 (54.3) | 23 (67.6) | 0.494 † | 47 (51.1) | 99 (54.1) | 0.83 † |
| Squamous carcinoma | 7 (20.0) | 6 (17.6) | 18 (19.6) | 31 (16.9) | ||
| Other | 9 (25.7) | 5 (14.7) | 27 (29.3) | 53 (29.0) | ||
| Stage | ||||||
| IIIB | 0 (0) | 5 (14.7) | 0.025 † | 8 (8.7) | 10 (5.5) | 0.312 † |
| IV | 35 (100) | 29 (85.3) | 84 (91.3) | 173 (94.5) | ||
| ERCC1 | ||||||
| Median | 33.4 | 36.7 | 0.72 * | 68.4 | 79.7 | 0.042 * |
| Range | 5.2–127.6 | 12.8–131.3 | 9.4–255.8 | 13.2–255.3 | ||
| RRM1 | ||||||
| Median | 39.1 | 32.9 | 0.146 * | 93 | 78.7 | 0.319 * |
| Range | 5.2–90.1 | 6.4–105.6 | 4.3–248.8 | 2.9–255.0 | ||
3.2. Testing Trial NCT00499109
| Variables | All Patients | p | Stage IV Patients | p | ||
|---|---|---|---|---|---|---|
| Low ERCC1 (n = 58) No (%) | High ERCC1 (n = 216) No (%) | Low ERCC1 (n = 54) No (%) | High ERCC1 (n = 202) No (%) | |||
| Age (years) | ||||||
| Median | 64.6 | 63.7 | 0.872 * | 64.6 | 63.8 | 0.924 * |
| Range | 42.6–85.0 | 39.6–83.8 | 42.6–85.0 | 39.6–83.8 | ||
| Sex | ||||||
| Male | 28 (48.3) | 105 (48.6) | 1.000 † | 25 (46.3) | 97 (48.0) | 0.879 † |
| Female | 30 (51.7) | 111 (51.4) | 29 (53.7) | 105 (52.0) | ||
| Histology | ||||||
| Adenocarcinoma | 34 (58.6) | 112 (51.9) | 0.607 † | 31 (57.4) | 107 (53.0) | 0.800 † |
| Squamous carcinoma | 8 (13.8) | 41 (19.0) | 8 (14.8) | 38 (18.8) | ||
| Other | 16 (27.6) | 63 (29.2) | 15 (27.8) | 57 (28.2) | ||
| Stage | ||||||
| IIIB | 4 (6.9) | 14 (6.5) | 1.00 † | 0 (0) | 0 (0) | NA |
| IV | 54 (93.1) | 202 (93.5) | 54 (100) | 202 (100) | ||
| ERCC1 | ||||||
| Median | 25.1 | 94.2 | <0.001 * | 25.1 | 94.7 | <0.001 * |
| Range | 9.4–38.4 | 39.4–255.8 | 9.4–38.4 | 39.4–255.8 | ||
| RRM1 | ||||||
| Median | 43.8 | 93.5 | <0.001 * | 44.8 | 92.9 | <0.001 * |
| Range | 2.9–233.9 | 7.7–255.0 | 2.9–233.9 | 7.7–255.0 | ||
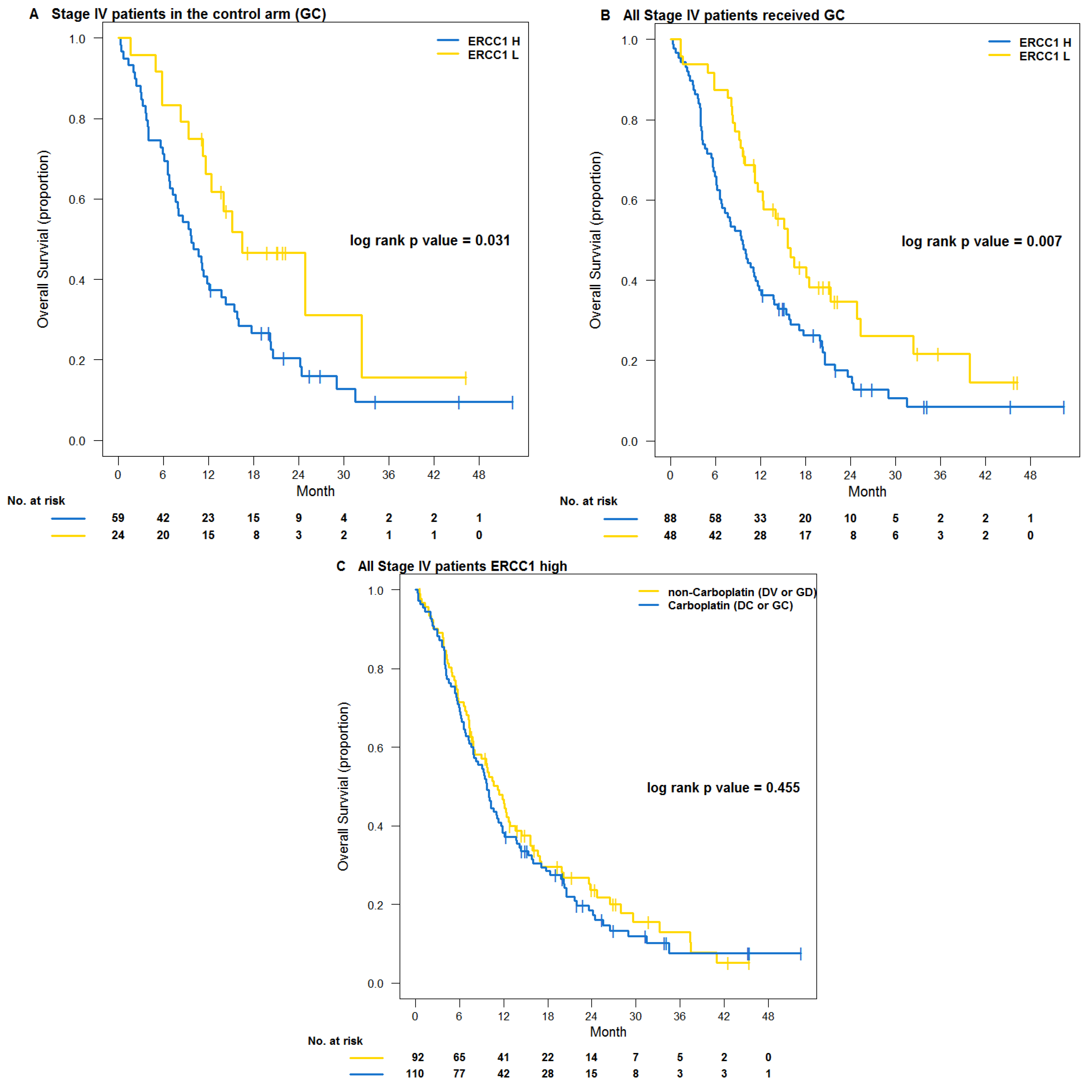
4. Discussions
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Cancer Facts and Figures; American Cancer Society: Atlanta, GA, USA, 2012.
- Hirsch, F.R.; Bunn, P.A. EGFR testing in lung cancer is ready for prime time. Lancet Oncol. 2009, 10, 432–433. [Google Scholar] [CrossRef]
- Kwak, E.L.; Bang, Y.J.; Camidge, D.R.; Shaw, A.T.; Solomon, B.; Maki, R.G.; Ou, S.H.I.; Dezube, B.J.; Janne, P.A.; Costa, D.B.; et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010, 363, 1693–1703. [Google Scholar]
- Zheng, Z.; Chen, T.; Li, X.; Haura, E.; Sharma, A.; Bepler, G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N. Engl. J. Med. 2007, 356, 800–808. [Google Scholar] [CrossRef]
- Olaussen, K.A.; Dunant, A.; Fouret, P.; Brambilla, E.; Andre, F.; Haddad, V.; Taranchon, E.; Filipits, M.; Pirker, R.; Popper, H.H.; et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N. Engl. J. Med. 2006, 355, 983–991. [Google Scholar] [CrossRef]
- Reynolds, C.; Obasaju, C.; Schell, M.J.; Li, X.; Zheng, Z.; Boulware, D.; Caton, J.R.; Demarco, L.C.; O’Rourke, M.A.; Wright, G.S.; et al. Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J. Clin. Oncol. 2009, 27, 5808–5815. [Google Scholar] [CrossRef]
- Soria, J.C. ERCC1-tailored chemotherapy in lung cancer: The first prospective randomized trial. J. Clin. Oncol. 2007, 25, 2648–2649. [Google Scholar] [CrossRef]
- Olaussen, K.A.; Mountzios, G.; Soria, J.C. ERCC1 as a risk stratifier in platinum-based chemotherapy for non-small-cell lung cancer. Curr. Opin. Pulm. Med. 2007, 13, 284–289. [Google Scholar] [CrossRef]
- Simon, G.R.; Schell, M.J.; Begum, M.; Kim, J.; Chiappori, A.; Haura, E.; Antonia, S.; Bepler, G. Preliminary indication of survival benefit from ERCC1 and RRM1-tailored chemotherapy in patients with advanced nonsmall cell lung cancer: Evidence from an individual patient analysis. Cancer 2012, 118, 2525–2531. [Google Scholar] [CrossRef]
- Filipits, M.; Pirker, R. Predictive markers in the adjuvant therapy of non-small cell lung cancer. Lung Cancer 2011, 74, 355–363. [Google Scholar] [CrossRef]
- Bepler, G.; Olaussen, K.A.; Vataire, A.L.; Soria, J.C.; Zheng, Z.; Dunant, A.; Pignon, J.P.; Schell, M.J.; Fouret, P.; Pirker, R.; et al. ERCC1 and RRM1 in the international adjuvant lung trial by automated quantitative in situ analysis. Am. J. Pathol. 2011, 178, 69–78. [Google Scholar] [CrossRef]
- Simon, G.R.; Sharma, S.; Cantor, A.; Smith, P.; Bepler, G. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest 2005, 127, 978–983. [Google Scholar] [CrossRef]
- Friboulet, L.; Olaussen, K.A.; Pignon, J.P.; Shepherd, F.A.; Tsao, M.S.; Graziano, S.; Kratzke, R.; Douillard, J.Y.; Seymour, L.; Pirker, R.; et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N. Engl. J. Med. 2013, 368, 1101–1110. [Google Scholar] [CrossRef]
- Bepler, G.; Williams, C.; Schell, M.J.; Chen, W.; Zheng, Z.; Simon, G.; Gadgeel, S.; Zhao, X.; Schreiber, F.; Brahmer, J.; et al. Randomized international Phase III trial of ERCC1 and RRM1 expression-based chemotherapy versus gemcitabine/carboplatin in advanced non-small-cell lung cancer. J. Clin. Oncol. 2013, 31, 2404–2412. [Google Scholar] [CrossRef]
- Camp, R.L.; Chung, G.G.; Rimm, D.L. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat. Med. 2002, 8, 1323–1327. [Google Scholar] [CrossRef]
- Otsuka, S.; Klimowicz, A.C.; Kopciuk, K.; Petrillo, S.K.; Konno, M.; Hao, D.; Muzik, H.; Stolte, E.; Boland, W.; Morris, D.; et al. CXCR4 overexpression is associated with poor outcome in females diagnosed with stage IV non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 1169–1178. [Google Scholar] [CrossRef]
- Mascaux, C.; Wynes, M.W.; Kato, Y.; Tran, C.; Asuncion, B.R.; Zhao, J.M.; Gustavson, M.; Ranger-Moore, J.; Gaire, F.; Matsubayashi, J.; et al. EGFR protein expression in non-small cell lung cancer predicts response to an EGFR tyrosine kinase inhibitor—A novel antibody for immunohistochemistry or AQUA technology. Clin. Cancer Res. 2011, 17, 7796–7807. [Google Scholar] [CrossRef]
- Dimou, A.; Agarwal, S.; Anagnostou, V.; Viray, H.; Christensen, S.; Rothberg, B.G.; Zolota, V.; Syrigos, K.; Rimm, D.L. Standardization of epidermal growth factor receptor (EGFR) measurement by quantitative immunofluorescence and impact on antibody-based mutation detection in non-small cell lung cancer. Am. J. Pathol. 2011, 179, 580–589. [Google Scholar] [CrossRef]
- Anagnostou, V.K.; Lowery, F.J.; Zolota, V.; Tzelepi, V.; Gopinath, A.; Liceaga, C.; Panagopoulos, N.; Frangia, K.; Tanoue, L.; Boffa, D.; et al. High expression of BCL-2 predicts favorable outcome in non-small cell lung cancer patients with non squamous histology. BMC Cancer 2010, 10, e186. [Google Scholar] [CrossRef]
- Anagnostou, V.K.; Bepler, G.; Syrigos, K.N.; Tanoue, L.; Gettinger, S.; Homer, R.J.; Boffa, D.; Detterbeck, F.; Rimm, D.L. High expression of mammalian target of rapamycin is associated with better outcome for patients with early stage lung adenocarcinoma. Clin. Cancer Res. 2009, 15, 4157–4164. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, X.; Schell, M.J.; Chen, T.; Boulware, D.; Robinson, L.; Sommers, E.; Bepler, G. Thymidylate synthase in situ protein expression and survival in Stage I non-small-cell lung cancer. Cancer 2008, 112, 2765–2773. [Google Scholar] [CrossRef]
- Chen, W.; Ghosh, D.; Raghunathan, T.E.; Norkin, M.; Sargent, D.J.; Bepler, G. On Bayesian methods of exploring qualitative interactions for targeted treatment. Stat. Med. 2012, 31, 3693–3707. [Google Scholar] [CrossRef]
- R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011.
- Lunn, D.J.; Thomas, A.; Best, N.; Spiegelhalter, D. WinBUGS—A Bayesian modelling framework: Concepts, structure, and extensibility. Stat. Comput. 2000, 10, 325–337. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chen, W.; Bepler, G. A Method for Biomarker Directed Survival Prediction in Advanced Non-Small-Cell Lung Cancer Patients Treated with Carboplatin-Based Therapy. J. Pers. Med. 2013, 3, 251-262. https://doi.org/10.3390/jpm3030251
Chen W, Bepler G. A Method for Biomarker Directed Survival Prediction in Advanced Non-Small-Cell Lung Cancer Patients Treated with Carboplatin-Based Therapy. Journal of Personalized Medicine. 2013; 3(3):251-262. https://doi.org/10.3390/jpm3030251
Chicago/Turabian StyleChen, Wei, and Gerold Bepler. 2013. "A Method for Biomarker Directed Survival Prediction in Advanced Non-Small-Cell Lung Cancer Patients Treated with Carboplatin-Based Therapy" Journal of Personalized Medicine 3, no. 3: 251-262. https://doi.org/10.3390/jpm3030251




