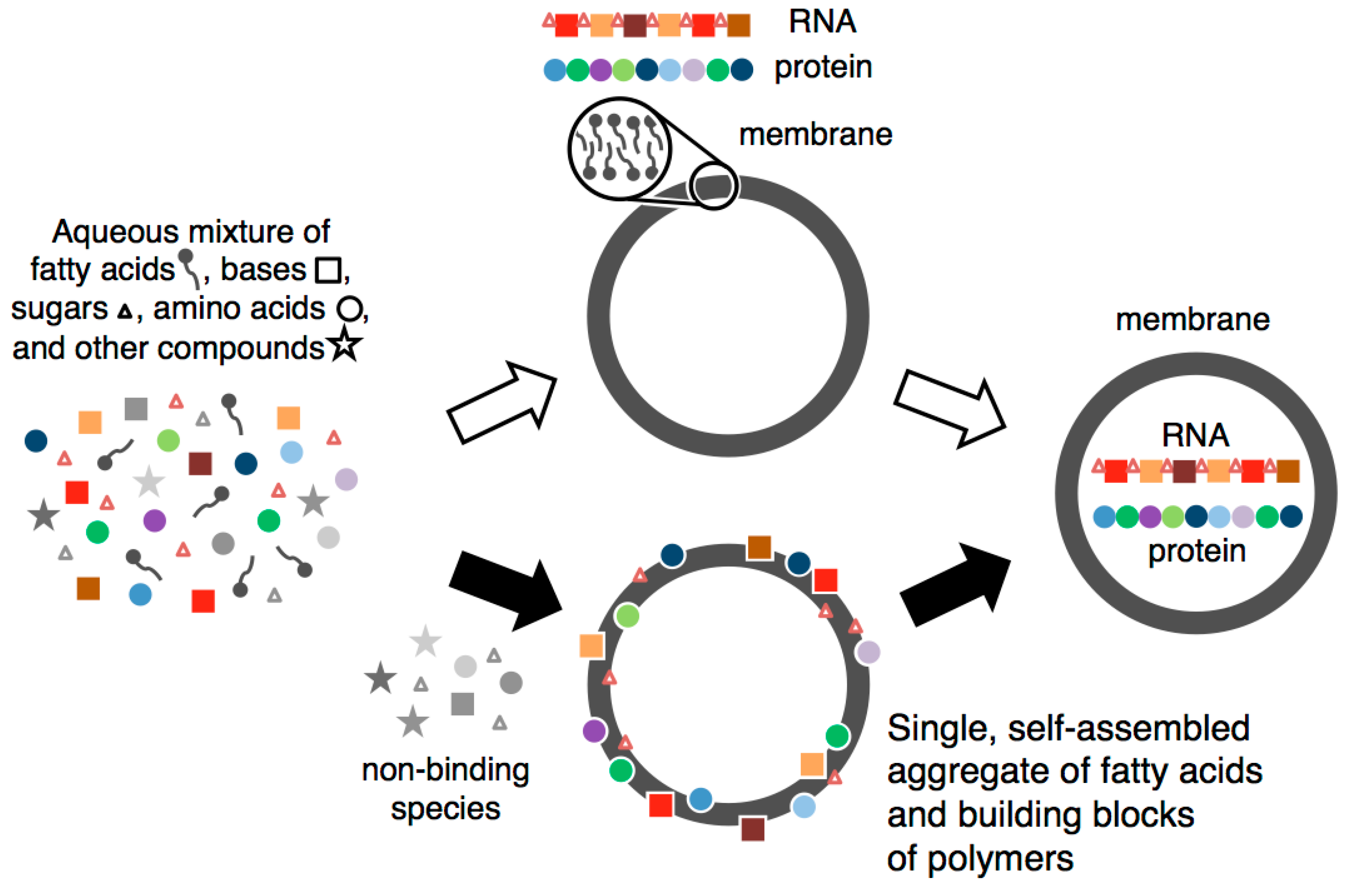
A Self-Assembled Aggregate Composed of a Fatty Acid Membrane and the Building Blocks of Biological Polymers Provides a First Step in the Emergence of Protocells
Abstract
:1. Introduction
- a.
- Fatty acids self-assembled in water to form a membrane.
- b.
- Components of RNA and protein were selected and concentrated via binding to self-assembled fatty acid membranes.
- c.
- These bound building blocks stabilized fatty acid membranes against salt-induced flocculation and increased the rate of vesicle formation.
- d.
- Membranes that were more stable bound more building blocks, leading to an auto-amplifying system.
- e.
- The resulting aggregate facilitated the formation of nucleosides, oligonucleotides and peptides, both because of the selection and concentration of building blocks and because of the conformational constraints and altered chemical environment due to binding.
- f.
- The oligomers, initially composed of random sequences, stabilized membranes and induced membrane growth more effectively than their unjoined components did, leading to the accumulation of oligonucleotides and peptides prior to the evolution of their complex functions in metabolism and information transfer.
2. Formation and Stabilization of a Self-Assembled Aggregate of Prebiotic Building Blocks
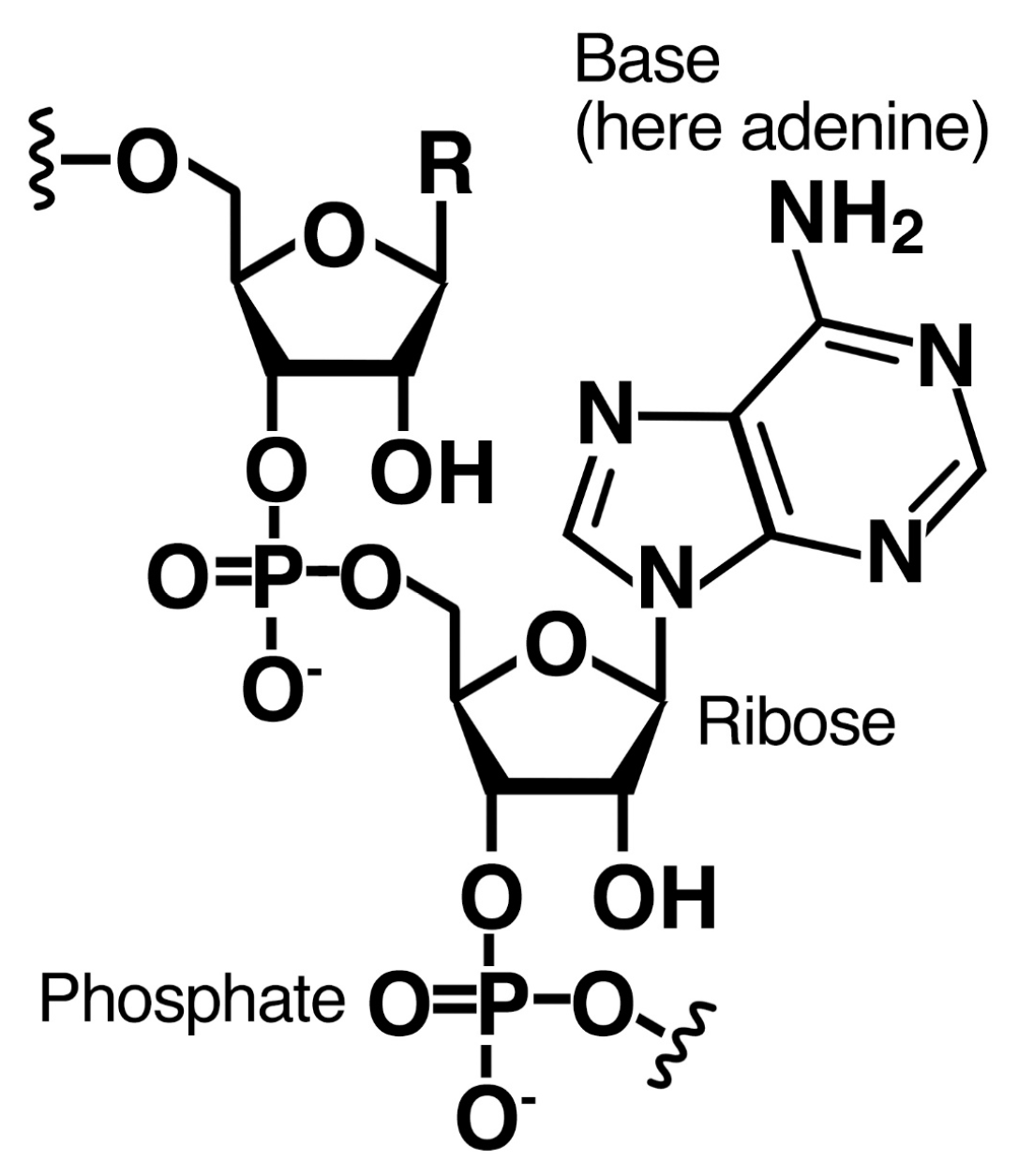
2.1. Amphiphiles
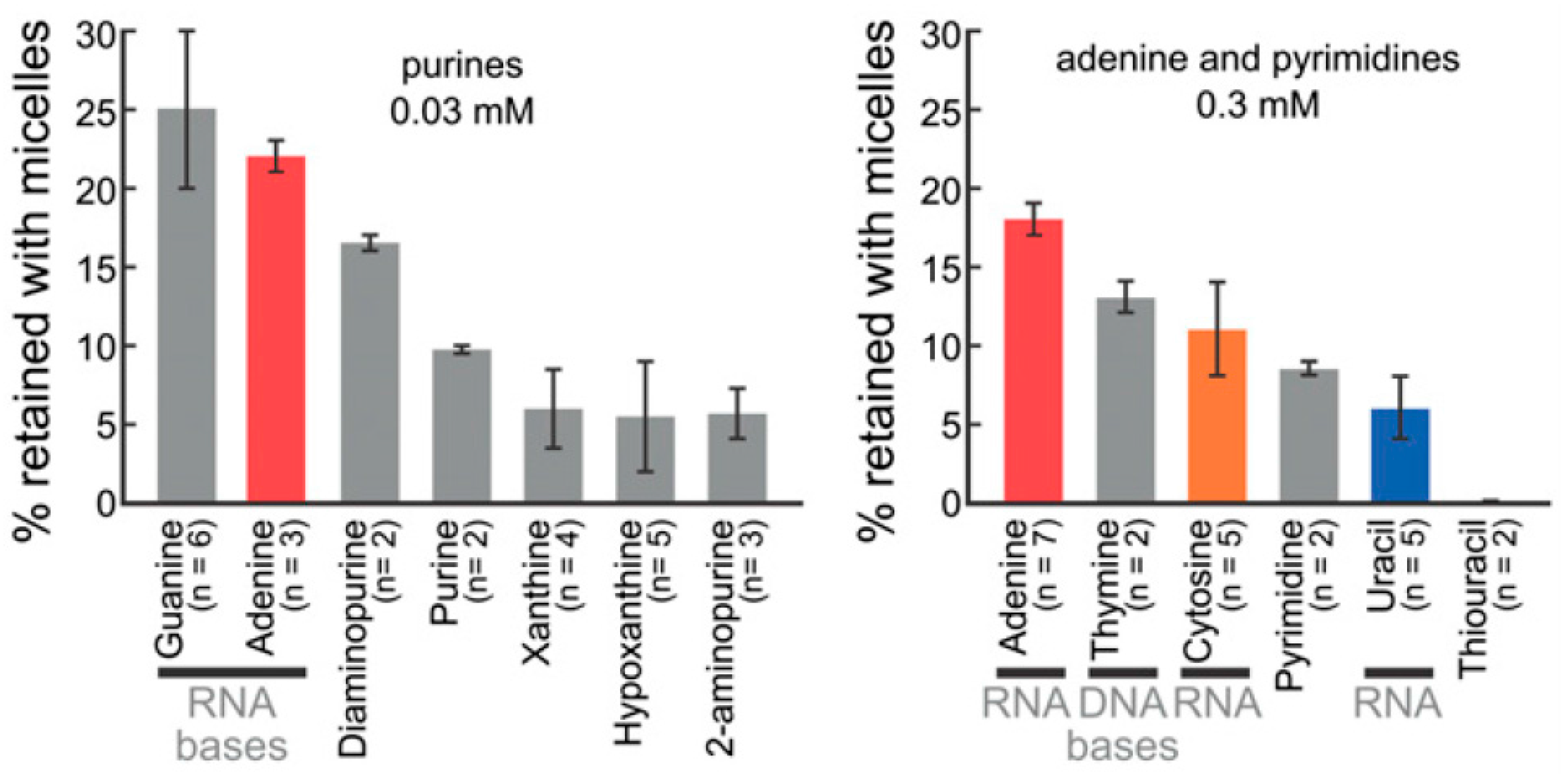
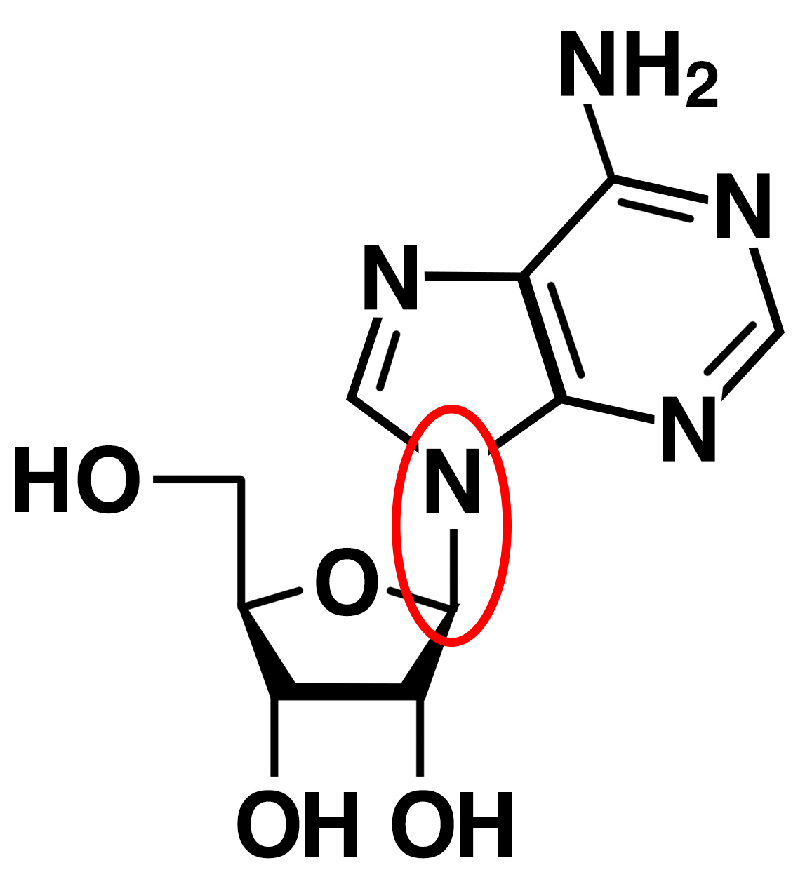
2.2. Bases
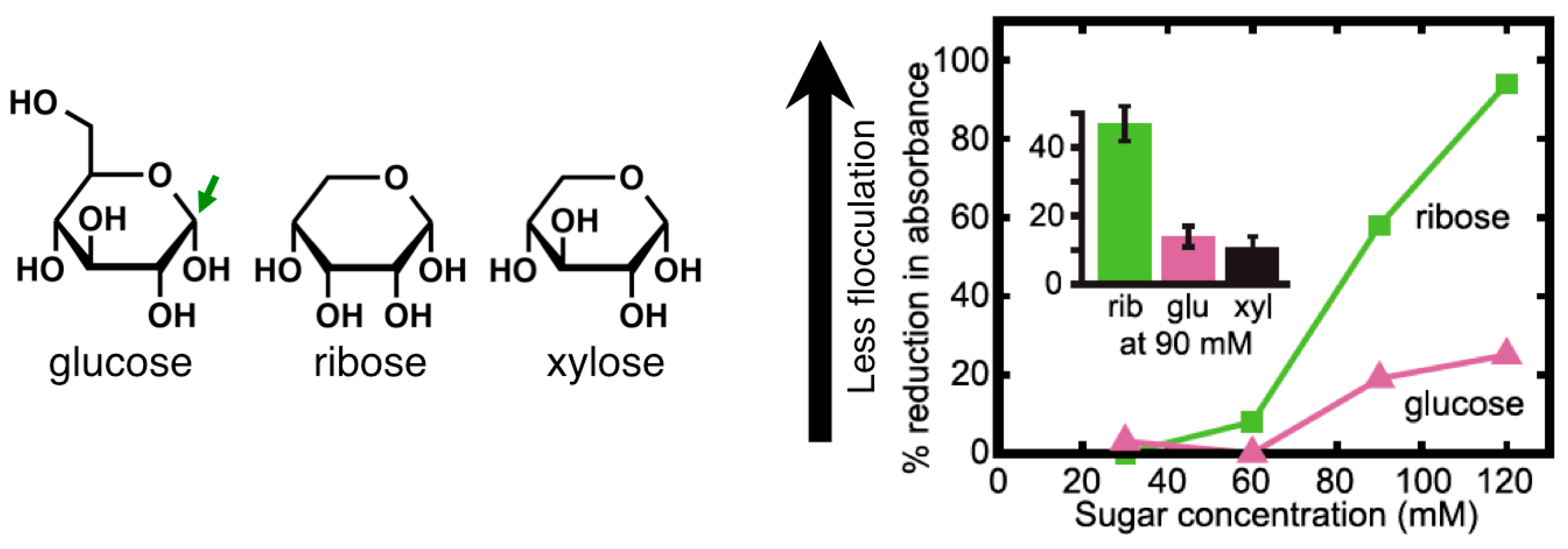
2.3. Sugars
2.4. Amino Acids and Dipeptides
2.5. Simultaneous Binding of Multiple RNA and Protein Building Blocks to Fatty Acid Membranes
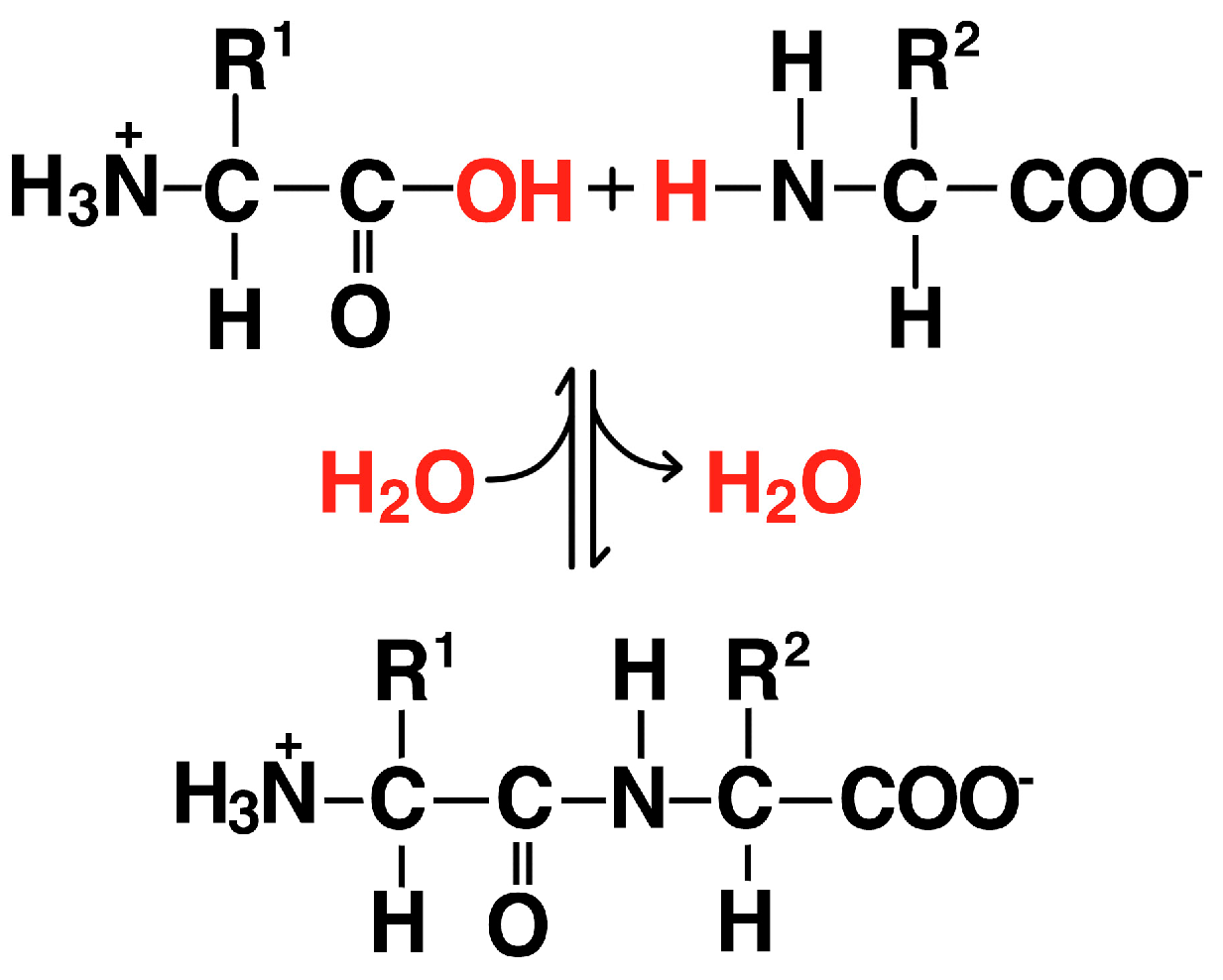
3. Formation of Oligomers
3.1. RNA
3.2. Proteins
4. From an Assemblage of the Components to an Evolving Protocell
- Some short oligomers catalyzed condensation (i.e., covalent linkage) between building blocks to form new oligomers, including at least some copies of itself.
- The presence of oligomers increased the stability, growth or division of vesicles to a greater extent than monomers.
- Vesicles bearing catalytic oligomers would have a higher overall concentration of oligomers, and would accumulate fatty acid and biopolymer building blocks at the expense of vesicles that lack catalytic oligomers.
- If the condensation mechanism joined short oligomers as well as single building blocks, longer oligomers would accumulate (assuming the reaction was faster than hydrolysis).
5. Conclusions
Acknowledgements
Conflicts of Interest
References
- Deamer, D. First Life; University of California Press: Berkeley, CA, USA, 2011; pp. 53–71. [Google Scholar]
- Black, R.A.; Blosser, M.C.; Stottrup, B.L.; Tavakley, R.; Deamer, D.W.; Keller, S.L. Nucleobases bind to and stabilize aggregates of a prebiotic amphiphile, providing a viable mechanism for the emergence of protocells. Proc. Natl. Acad. Sci. USA 2013, 110, 13272–13276. [Google Scholar] [CrossRef] [PubMed]
- Hud, N.V.; Cafferty, B.J.; Krishnamurthy, R.; Williams, L.D. The origin of RNA and “my grandfather’s axe”. Chem. Biol. 2013, 20, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, R. On the emergence of RNA. Isr. J. Chem. 2015. [Google Scholar] [CrossRef]
- Fry, I. The Emergence of Life on Earth: A Historical and Scientific Overview; Rutgers University Press: New Brunswick, NJ, USA, 2000; pp. 172–178. [Google Scholar]
- Luisi, P.L. The Emergence of Life; Cambridge University Press: Cambridge, UK, 2006; pp. 29–32. [Google Scholar]
- Woolf, N.J. A hypothesis about the origin of biology. Orig. Life Evol. Biosph. 2015, 45, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M.; Fujikawa, S.M.; Szostak, J.W. Experimental models of primitive cellular compartments: Encapsulation, growth, and division. Science 2003, 302, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M.; Mansy, S.S.; Szostak, J.W. Mineral surface directed membrane assembly. Orig. Life Evol. Biosph. 2007, 37, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Prebiotic systems chemistry: New perspectives for the origins of life. Chem. Rev. 2014, 114, 285–366. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.H.; Percivalle, C.; Ritson, D.J.; Duffy, C.D.; Sutherland, J.D. Common origins of RNA, protein, and lipid precursors in a cyanosolfidic protometabolism. Nat. Chem. 2015, 7, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Rajamani, S.; Vlassov, A.; Benner, S.; Coombs, A.; Olasagasti, F.; Deamer, D. Lipid-assisted synthesis of RNA-like polymers from mononucleotides. Orig. Life Evol. Biosph. 2008, 38, 57–74. [Google Scholar] [CrossRef] [PubMed]
- DeGuzman, V.; Vercoutere, W.; Shenasa, H.; Deamer, D. Generation of oligonucleotides under hydrothermal conditions by non-enzymatic polymerization. J. Mol. Evol. 2014, 78, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Damer, B.; Deamer, D. Coupled phases and combinatorial selection in fluctuating hydrothermal pools: A scenario to guide experimental approaches to the origin of cellular life. Life 2015, 5, 872–887. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.A.; Roberts, R.W.; Szostak, J.W. The emergence of competition between model protocells. Science 2004, 305, 1474–1476. [Google Scholar] [CrossRef] [PubMed]
- Deamer, D.; Dworkin, J.P.; Sandford, S.A.; Bernstein, M.P.; Allamandola, L.J. The first cell membranes. Astrobiology 2002, 2, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Morigaki, K.; Walde, P. Fatty acid vesicles. Curr. Opin. Coll. Int. Sci. 2007, 12, 75–80. [Google Scholar] [CrossRef]
- Apel, C.L.; Deamer, D.W.; Mauntner, M.N. Self-assembled vesicles of monocarboxylic acids and alcohols: Conditions for stability and for the encapsulation of biopolymers. Biochim. Biophys. Acta 2002, 1559, 1–9. [Google Scholar] [CrossRef]
- Proskurowski, G.; Lilley, M.D.; Seewald, J.S.; Früh-Green, G.L.; Olson, E.J.; Lupton, J.E.; Sylva, S.P.; Kelley, D.S. Abiogenic hydrocarbon production at Lost City hydrothermal field. Science 2008, 319, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Lawless, J.; Yuen, G. Quantification of monocarboxylic acids in the Murchison carbonaceous meteorite. Nature 1979, 282, 396–398. [Google Scholar] [CrossRef]
- Berclaz, N.; Muller, M.; Walde, P.; Luisi, P.L. Growth and transformation of vesicles studied by ferritin labeling and cryotransmission electron microscopy. J. Phys. Chem. B 2001, 105, 1056–1064. [Google Scholar] [CrossRef]
- Walde, P.; Wick, R.; Fresta, M.; Mangone, A.; Luisi, P.L. Autopoietic self-reproduction of fatty acid vesicles. J. Am. Chem. Soc. 1994, 116, 11649–11654. [Google Scholar] [CrossRef]
- Naraoka, H.; Shimoyama, A.; Harada, K. Molecular distribution of monocarboxylic acids in Asuka carbonaceous chondrites from Antarctica. Orig. Life Evol. Biosph. 1999, 29, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Budin, I.; Prywes, N.; Zhang, N.; Szostak, J.W. Chain-length heterogeneity allows for the assembly of fatty acid vesicles in dilute solutions. Biophys. J. 2014, 107, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Namani, T.; Walde, P. From decanoate micelles to decanoic acid/dodecylbenzenesulfonate vesicles. Langmuir 2005, 21, 6210–6219. [Google Scholar] [CrossRef] [PubMed]
- Monnard, P.-A.; Apel, C.; Kanavarioti, A.; Deamer, D. Influence of ionic inorganic solutes on self-assembly and polymerization processes related to early forms of life: Implications for a prebiotic aqueous medium. Astrobiology 2002, 2, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.A.; Salehi-Ashtiani, K.; Szostak, J.W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005, 127, 13351–13355. [Google Scholar] [CrossRef] [PubMed]
- Maurer, S.; Deamer, D.; Boncella, J.; Monnard, P.-A. Chemical evolution of amphiphiles: Glycerol monoacyl derivatives stabilize plausible prebiotic membranes. Astrobiology 2009, 9, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Callahan, M.P.; Smith, K.E.; Cleaves, H.J.; Ruzicka, J.; Stern, J.C.; Glavin, D.P.; House, C.H.; Dworkin, J.P. Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases. Proc. Natl. Acad. Sci. USA 2011, 108, 13995–13998. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, R. The role of pKa of nucleobases on the orgins of chemical evolution. Acc. Chem. Res. 2012, 45, 2035–2044. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.D.; Chawla, B.; Shaw, B.R. The hydrogen bonding of cytosine and guanine: Calorimetric and 1H-NMR analysis of the molecular interactions of nucleic acid bases. Biopolymers 1987, 26, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Ricardo, A.; Carrigan, M.A.; Olcott, A.N.; Benner, S.A. Borate minerals stabilize ribose. Science 2004, 303, 196. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, Y.; Horiuchi, M.; Kakegawa, T. Selective stabilization of ribose by borate. Orig. Life Evol. Biosph. 2013, 43, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Meinert, C.; Myrgorodska, J.; de Marcellus, P.; Buhse, T.; Nahon, L.; Hoffman, S.V.; le Sergeant d’Hendecourt, L.; Meierhenrich, U.J. Ribose and related sugars from ultraviolet irradiation of interstellar ice analogs. Science 2016, 352, 208–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansy, S.S.; Szostak, J.W. Thermostability of model protocell membranes. Proc. Natl. Acad. Sci. USA 2008, 105, 13351–13355. [Google Scholar] [CrossRef] [PubMed]
- Longo, L.M.; Lee, J.; Blaber, M. Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein. Proc. Natl. Acad. Sci. USA 2013, 110, 2135–2139. [Google Scholar] [CrossRef] [PubMed]
- Raggi, L.; Bada, J.L.; Lazcano, A. On the lack of evolutionary continuity between prebiotic peptides and extant enzymes. Phys. Chem. Chem. Phys. 2016, 18, 20028–20032. [Google Scholar] [CrossRef] [PubMed]
- Parker, E.T.; Zhou, M.; Burton, A.S.; Glavin, D.P.; Dworkin, J.P.; Krishnamurthy, R.; Fernandez, F.M.; Bada, J.L. A plausible simultaneous synthesis of amino acids and simple peptides on the primordial earth. Angew. Chem. 2014, 126, 8270–8274. [Google Scholar] [CrossRef]
- Shimoyama, A.; Ogasawara, R. Dipeptides and diketopiperazines in the Yamato-791198 and Murchison carbonaceous chondrites. Orig. Life Evol. Biosph. 2002, 32, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Namani, T.; Deamer, D. Stability of model membranes in extreme environments. Orig. Life Evol. Biosph. 2008, 38, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Novales, B.; Riaublanc, A.; Navailles, L.; Houssou, B.; Gaillard, C.; Nallet, F.; Douliez, J.-P. Self-assembly and foaming properties of fatty acid-lysine aqueous dispersions. Langmuir 2010, 26, 5329–5334. [Google Scholar] [CrossRef] [PubMed]
- Adamala, K.; Szostak, J.W. Competition between model protocells driven by an encapsulated catalyst. Nat. Chem. 2013, 5, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Murillo-Sanchez, S.; Beaufils, D.; Gonzalez Manas, J.M.; Pascal, R.; Ruiz-Mirazo, K. Fatty acids’ double role in the prebiotic formation of a hydrophobic dipeptide. Chem. Sci. 2016, 7, 3406–3413. [Google Scholar] [CrossRef]
- Kamat, N.P.; Tobe, S.; Hill, I.T.; Szostak, J.W. Electrostatic localization of RNA to protocell membranes by cationic hydrophobic peptides. Angew. Chem. Int. Ed. 2015, 127, 11901–11905. [Google Scholar] [CrossRef]
- Blocher, M.; Liu, D.; Walde, P.; Luisi, P.L. Liposome-assisted selective polycondensation of alpha-amino acids and peptides. Macromolecules 1999, 32, 7332–7334. [Google Scholar] [CrossRef]
- Blocher, M.; Liu, D.; Luisi, P.L. Liposome-assisted selective polycondensation of alpha-amino acids and peptides: The case of charged liposomes. Macromolecules 2000, 33, 5787–5796. [Google Scholar] [CrossRef]
- Pohorille, A.; Wilson, M.A.; Chipot, C. Membrane peptides and their role in protobiological evolution. Orig. Life Evol. Biosph. 2003, 33, 173–197. [Google Scholar] [CrossRef] [PubMed]
- Fallah-Araghi, A.; Meguellati, K.; Baret, J.-C.; El Harrak, A.; Mangeat, T.; Karplus, M.; Ladame, S.; Marques, C.M.; Griffiths, A.D. Enhanced chemical synthesis at soft interfaces: A universal reaction-adsorption mechanism in microcompartments. Phys. Rev. Lett. 2014, 112, 028301. [Google Scholar] [CrossRef] [PubMed]
- Pizzarello, S.; Weber, A.L. Stereoselective syntheses of pentose sugars under realistic prebiotic conditions. Orig. Life Evol. Biosph. 2010, 40, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M. Specific aminoacylation of C4N hairpin RNAs with the cognate aminoacyl-adenylates in the presence of a dipeptide: Origin of the genetic code. J. Biochem. 1995, 117, 23–26. [Google Scholar] [PubMed]
- Grochmal, A.; Prout, L.; Makin-Taylor, R.; Prohens, R.; Tomas, S. Modulation of reactivity in the cavity of liposomes promotes the formation of peptide bonds. J. Am. Chem. Soc. 2015, 137, 12269–12275. [Google Scholar] [CrossRef] [PubMed]
- Monnard, P.-A.; Walde, P. Current ideas about prebiological compartmentalization. Life 2015, 5, 1239–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuller, W.D.; Sanchez, R.A.; Orgel, L.E. Studies in prebiotic synthesis. VII: Solid-state synthesis of purine nucleosides. J. Mol. Evol. 1972, 1, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.D. Ribonucleotides. Cold Spring Harb. Perspect. Biol. 2010, 2, a005439. [Google Scholar] [CrossRef] [PubMed]
- Cafferty, B.J.; Hud, N.V. Abiotic synthesis of RNA in water: A common goal of prebiotic chemistry and bottom-up synthetic biology. Curr. Opin. Chem. Biol. 2014, 22, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Powner, M.W.; Gerland, B.; Sutherland, J.D. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 2009, 459, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.; Thoma, I.; Deutsch, A.; Gerhrke, T.; Mayer, P.; Zipse, H.; Carell, T. A high-yielding, strictly regioselective prebiotic purine nucleoside formation pathway. Science 2016, 352, 833–836. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A.; Kim, H.-J.; Carrigan, M.A. Asphalt, water, and the prebiotic synthesis of ribose, ribonucleotides, and RNA. Acc. Chem. Res. 2012, 45, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.C.; Cafferty, B.J.; Mamajanov, I.; Gallego, I.; Khanam, J.; Krishnamurthy, R.; Hud, N.V. Spontaneous prebiotic formation of a beta-ribofuranoside that self-assembles with a complementary heterocycle. J. Am. Chem. Soc. 2014, 136, 5640–5646. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Benner, S.A. Prebiotic glycosylation of uracil with electron-donating substituents. Astrobiology 2015, 15, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Cafferty, B.J.; Fialho, D.M.; Khanam, J.; Krishnamurthy, R.; Hud, N.V. Spontaneous formation and base pairing of plausible prebiotic nucleotides in water. Nat. Commun. 2016, 7, 11328. [Google Scholar] [CrossRef] [PubMed]
- Mungi, C.V.; Singh, S.K.; Chugh, J.; Rajamani, S. Synthesis of barbituric acid containing nucleotides and their implications for the origin of primitive informational polymers. Phys. Chem. Chem. Phys. 2016, 18, 20144–20152. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.P.; Joyce, G.F. The origins of the RNA world. Cold Spring Harb. Perspect. Biol. 2012, 4, a003608. [Google Scholar] [CrossRef] [PubMed]
- Pasek, M.A.; Harnmeijerb, J.P.; Buick, R.; Gull, G.; Atlas, Z. Evidence for reactive reduced phosphorus species in the early Archean ocean. Proc. Natl. Acad. Sci. USA 2013, 110, 10089–10094. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P.; Hill, A.R.; Liu, R.; Orgel, L.E. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 1996, 381, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.; Maurel, M.-C.; Deamer, D. Salt-promoted synthesis of RNA-like molecules in simulated hydrothermal conditions. J. Mol. Evol. 2014, 80, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Toppozini, L.; Dies, H.; Deamer, D.W.; Rheinstadter, M.C. Adenosine monophosphate forms ordered arrays in multilamellar lipid matrices: Insights into assembly of nucleic acid for primitive life. PLoS ONE 2013, 8, e62810. [Google Scholar] [CrossRef] [PubMed]
- Mungi, C.V.; Rajamani, S. Characterization of RNA-like oligomers from lipid-assisted nonenzymatic synthesis: implications for origin of informational molecules on early earth. Life 2015, 5, 65–84. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, C.; Liang, Y. FT-SERS studies on molecular recognition capabilities of monolayers of novel nucleolipid amphiphiles. Langmuir 2000, 16, 3937–3940. [Google Scholar] [CrossRef]
- Rosemeyer, H. Nucleolipids: Natural occurrence, synthesis, molecular recognition, and supramolecular assemblies as potential precursors of life and bioorganic materials. Chem. Biodivers. 2005, 2, 977–1063. [Google Scholar] [CrossRef] [PubMed]
- Rode, B.M. Peptides and the origin of life. Peptides 1999, 20, 773–786. [Google Scholar] [CrossRef]
- Leman, L.; Orgel, L.; Ghadiri, M.R. Carbonyl sulfide-mediated prebiotic formation of peptides. Science 2004, 306, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Danger, G.; Plasson, R.; Pascal, R. Pathways for the formation and evolution of peptides in prebiotic environments. Chem. Soc. Rev. 2012, 41, 5416–5429. [Google Scholar] [CrossRef] [PubMed]
- Cleaves, H.J.; Aubrey, A.D.; Bada, J.L. An evaluation of the critical parameters for abiotic peptide synthesis in submarine hydrothermal systems. Orig. Life Evol. Biosph. 2009, 39, 109–126. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, J.G.; Yu, S.-S.; Mamajanov, I.; Grover, M.A.; Krishnamurthy, R.; Fernandez, F.M.; Hud, N.V. Ester-mediated amide bond formation driven by wet-dry cycles: A possible path to polypeptides on the prebiotic earth. Angew. Chem. Int. Ed. 2015, 54, 9871–9875. [Google Scholar] [CrossRef] [PubMed]
- Zepik, H.H.; Rajamani, S.; Maurel, M.-C.; Deamer, D. Oligomerization of thioglutamic acid: Encapsulated reactions and lipid catalysis. Orig. Life Evol. Biosph. 2007, 37, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Griffith, E.C.; Vaida, V. In situ observation of peptide bond formation at the water-air interface. Proc. Natl. Acad. Sci. USA 2012, 109, 15697–15701. [Google Scholar] [CrossRef] [PubMed]
- Furuuchi, R.; Imai, E.-I.; Honda, H.; Hatori, K.; Matsuno, K. Evolving lipid vesicles in prebiotic hydrothermal environments. Orig. Life Evol. Biosph. 2005, 35, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.F.; Szostak, J.W. Coupled growth and division of model protocell membranes. J. Am. Chem. Soc. 2009, 131, 5705–5713. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.F.; Adamala, K.; Zhang, N.; Szostak, J.W. Photochemically driven redox chemistry induces protocell membrane pearling and division. Proc. Natl. Acad. Sci. USA 2012, 109, 9828–9832. [Google Scholar] [CrossRef] [PubMed]
- Adamala, K.P.; Engelhart, A.E.; Szostak, J.W. Collaboration between primitive cell membranes and soluble catalysts. Nat. Commun. 2016, 7, 11041. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.A.; Huynh, C.; Higgs, P.G. The origin and spread of a cooperative replicase in a prebiotic chemical system. J. Theor. Biol. 2015, 364, 249–259. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Black, R.A.; Blosser, M.C. A Self-Assembled Aggregate Composed of a Fatty Acid Membrane and the Building Blocks of Biological Polymers Provides a First Step in the Emergence of Protocells. Life 2016, 6, 33. https://doi.org/10.3390/life6030033
Black RA, Blosser MC. A Self-Assembled Aggregate Composed of a Fatty Acid Membrane and the Building Blocks of Biological Polymers Provides a First Step in the Emergence of Protocells. Life. 2016; 6(3):33. https://doi.org/10.3390/life6030033
Chicago/Turabian StyleBlack, Roy A., and Matthew C. Blosser. 2016. "A Self-Assembled Aggregate Composed of a Fatty Acid Membrane and the Building Blocks of Biological Polymers Provides a First Step in the Emergence of Protocells" Life 6, no. 3: 33. https://doi.org/10.3390/life6030033






