Evolutionary Aspects and Regulation of Tetrapyrrole Biosynthesis in Cyanobacteria under Aerobic and Anaerobic Environments
Abstract
:1. Introduction
2. Coexistence of Aerobic and Anaerobic Enzymes
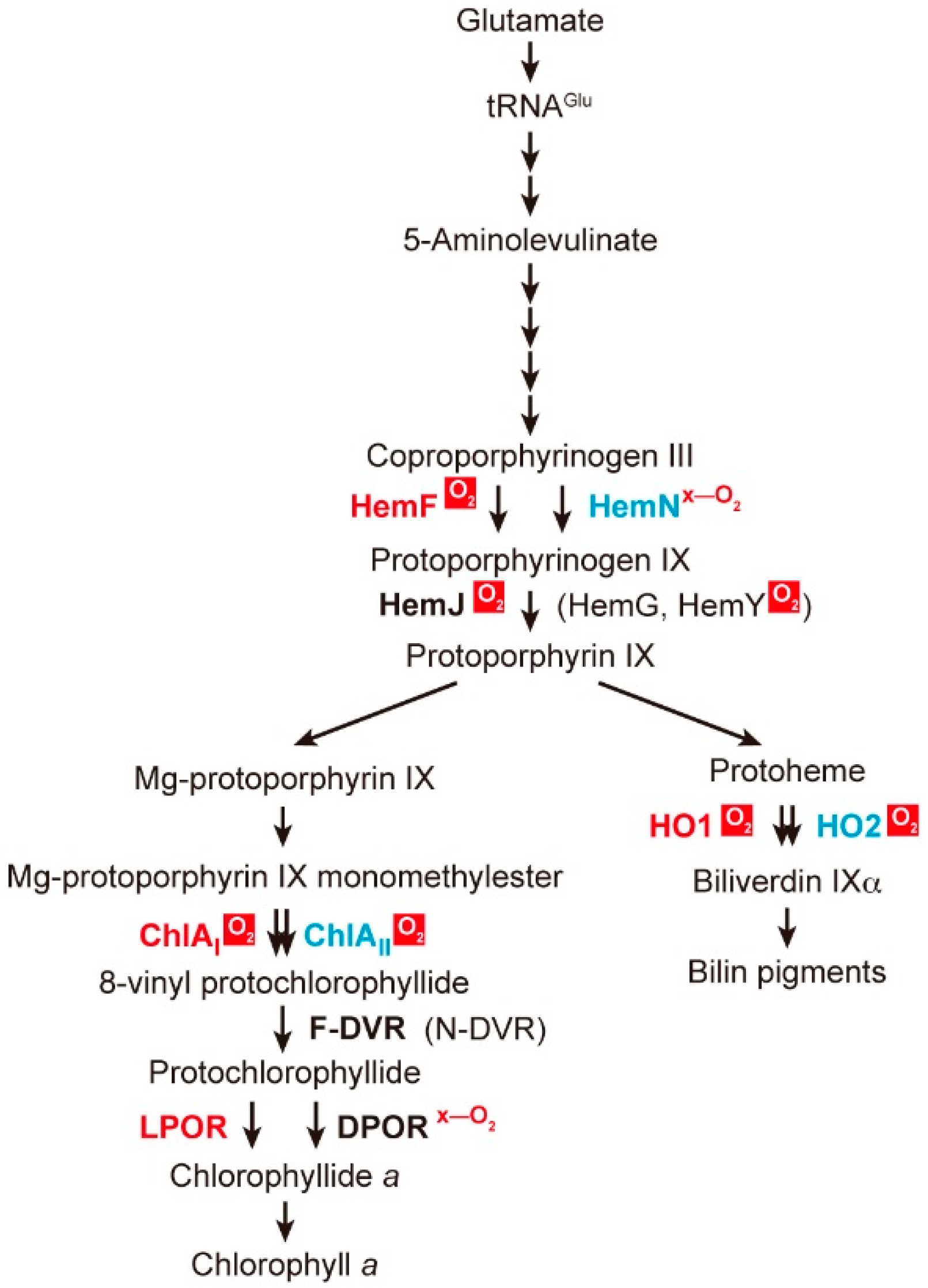
2.1. Coproporphyrinogen III Oxidation
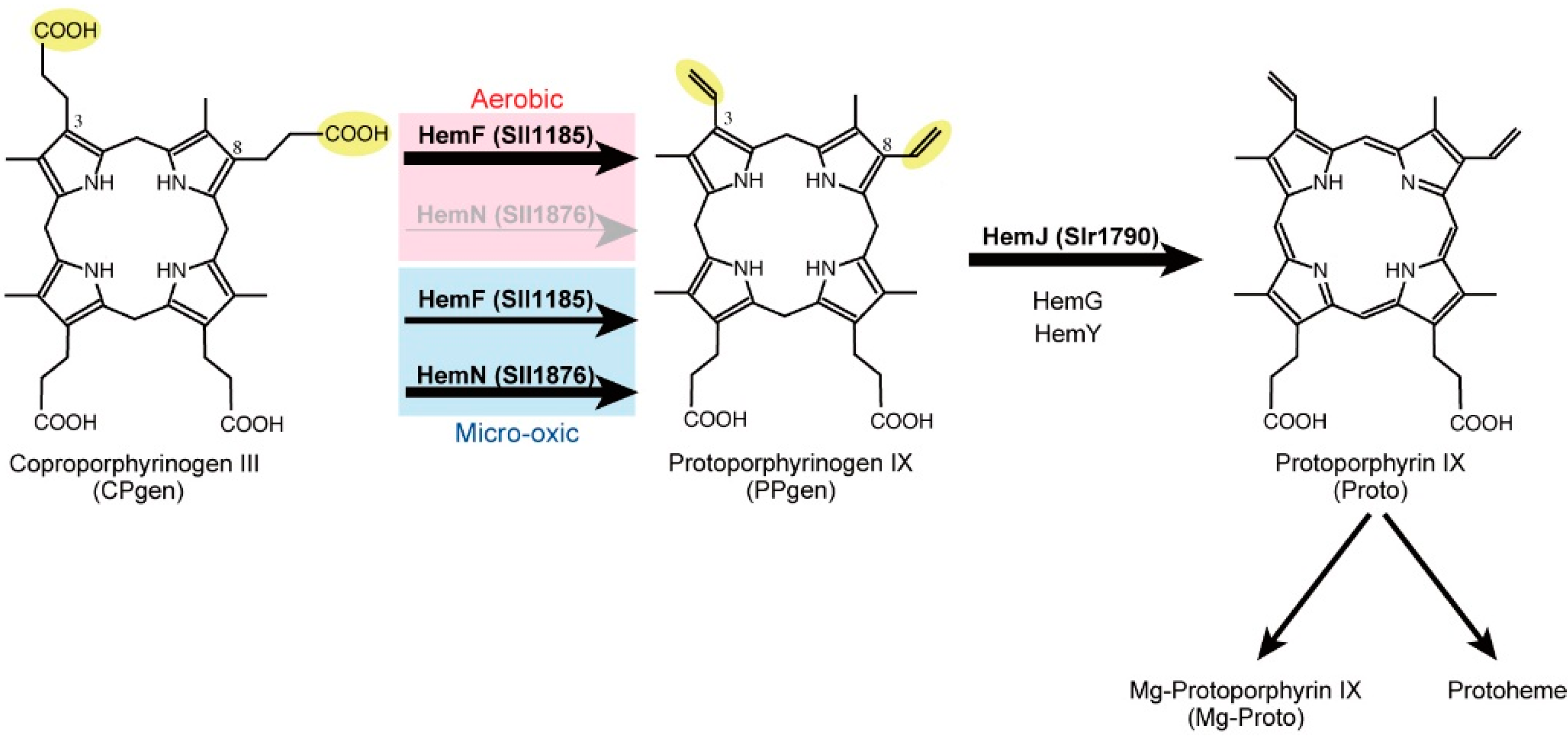
2.2. Protoporphyrinogen IX Oxidation
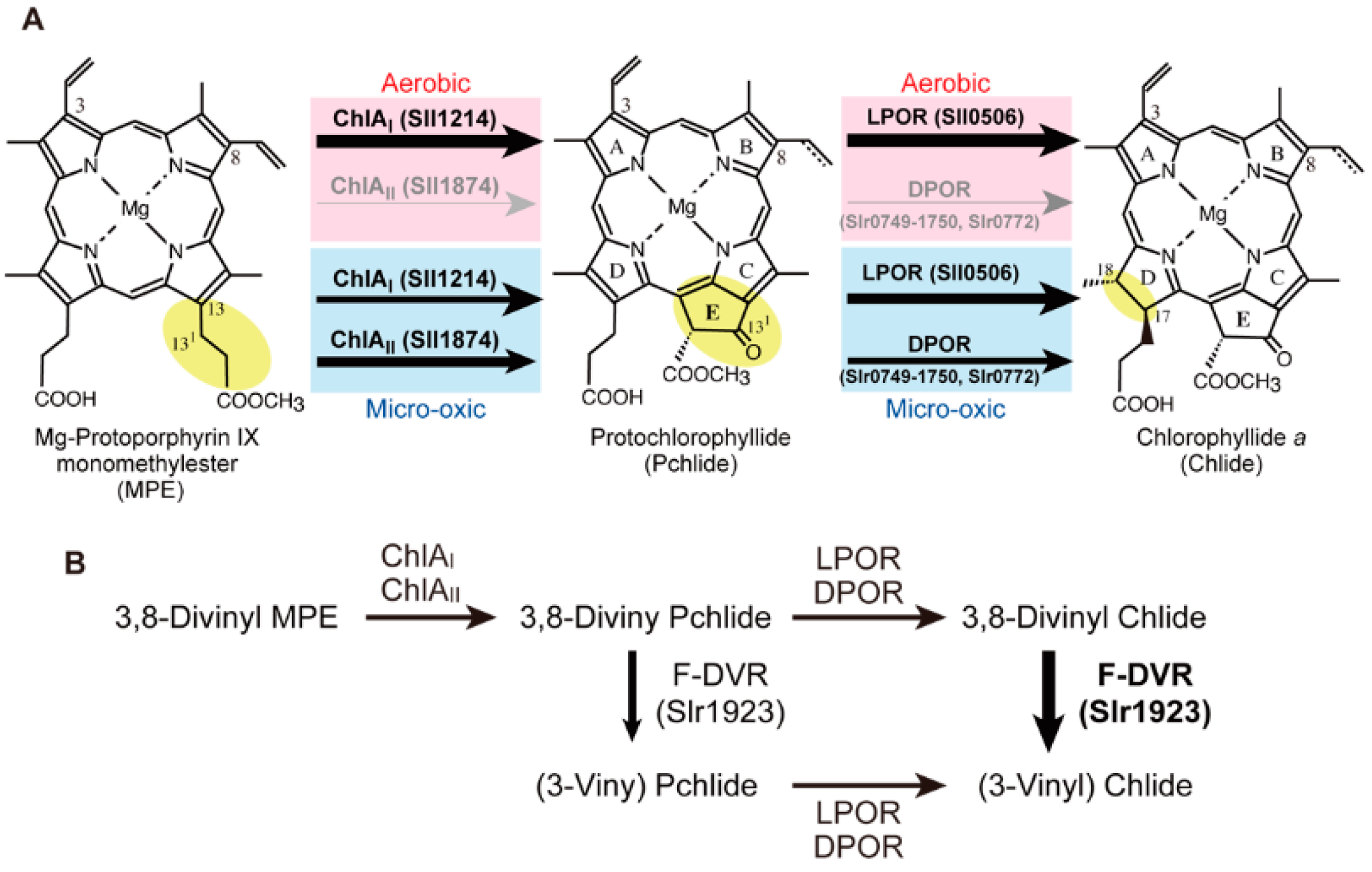
2.3. Mg-Protoporphyrin IX Monomethyl Ester Cyclization
| F/M a | Species b | chlR c | Cpgen Oxidation | MPE Cyclization | Heme Cleavage | Nif d | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| hemF | hemN | chlAI | chlAII | bchE | ho1 | ho2 | ||||
| F | Synechococcus elongatus PCC 6301 | + | + | – | + | – | – | + | – | – |
| F | Synechococcus elongatus PCC 7942 | + | + | – | + | – | – | + | – | – |
| F | Nostoc punctiforme | + | + | + ¶ | + | + | – | + | + ¶ | + |
| F | Anabaena sp. PCC 7120 | + | + §,e | + ¶ | + | + §,e | – | + | + ¶ | + |
| F | Anabaena variabilis | + | + | + ¶ | + | + | – | + | + ¶ | + |
| M | Trichodesmium erythraeum | + * | + | + | + | – | – | + | – | + |
| F | Cyanothece sp. PCC 7424 | + | + | + ¶ | + | + ¶ | – | + | + ¶ | + |
| F | Cyanothece sp. PCC 7822 | + | + | + ¶ | + | + ¶ | + | + | + ¶ | + |
| M | Cyanothece sp. ATCC 51142 | – | + | + ¶ | + | + | – | + | + ¶ | + |
| F | Cyanothece sp. PCC 8801 | + | + | + ¶ | + | + | – | + | + ¶ | + |
| F | Cyanothece sp. PCC 8802 | + | + | + ¶ | + | + | – | + | + ¶ | + |
| F | Microcystis aeruginosa | – | + | – | + | – | – | + | – | – |
| F | Synechocystis sp. PCC 6803 | + * | + § | + ¶ | + | + ¶ | – | + § | + ¶ | – |
| F | Leptolyngbya boryana | + * | + | + ¶ | + | + ¶ | – | + | + ¶ | + |
| M | Synechococcus sp. PCC 7002 | + * | + | + ¶ | + | + ¶ | – | + | + ¶ | – |
| M | Acaryochloris marina | + | + | + ¶ | + | + ¶ | – | + | + ¶ | – |
| F | Cyanothece sp. PCC 7425 | + | + | + ¶ | + | + ¶ | + ¶ | + | + ¶ | + |
| F | Thermosynechococcus elongatus BP-1 | – | + | + ¶ | + | + ¶ | – | + | + ¶ | – |
| F | Cyanobacteria Yellowstone A-Prime | – | + | – | + | – | – | + | – | + |
| F | Cyanobacteria Yellowstone B-Prime | – | + | – | + | – | – | + | – | + |
| F | Gloeobacter violaceus PCC 7421 | + | + | – | + | – | – | + | – | – |
| M | Prochlorococcus marinus MIT 9301 | – | + | – | + | – | – | + | – | – |
| M | Prochlorococcus marinus AS9601 | – | + | – | + | – | – | + | – | – |
| M | Prochlorococcus marinus MIT 9312 | – | + | – | + | – | – | + | – | – |
| M | Prochlorococcus marinus MIT 9515 | – | + | – | + | – | – | + | – | – |
| M | Prochlorococcus marinus MED4 | – | + | – | + | – | – | + | – | – |
| M | Prochlorococcus marinus NATL2A | – | + | – | – | + | – | + | – | – |
| M | Prochlorococcus marinus NATL1A | – | + | – | – | + | – | + | – | – |
| M | Prochlorococcus marinus SS120 | – | + | – | – | + | – | + | – | – |
| M | Prochlorococcus marinus MIT 9313 | – | + | – | – | + | – | + | – | – |
| M | Prochlorococcus marinus MIT 9303 | – | + | – | + f | – | – | – | – | |
| M | Synechococcus sp. WH 7803 | – | + | – | + | – | – | + | – | – |
| M | Synechococcus sp. CC 9311 | – | + | – | + | – | – | + | – | – |
| M | Synechococcus sp. WH 8102 | + | + | – | + | – | – | + | – | – |
| M | Synechococcus sp. RCC 307 | – | + | – | + | – | – | + | – | – |
| M | Synechococcus sp. CC 9902 | – | + | – | + | – | – | + | – | – |
| M | Synechococcus sp. CC 9605 | – | + | – | + | – | – | + | – | – |
2.4. Protochlorophyllide Reduction
2.5. 8-Vinyl Reduction
2.6. Heme Oxygenase Reaction
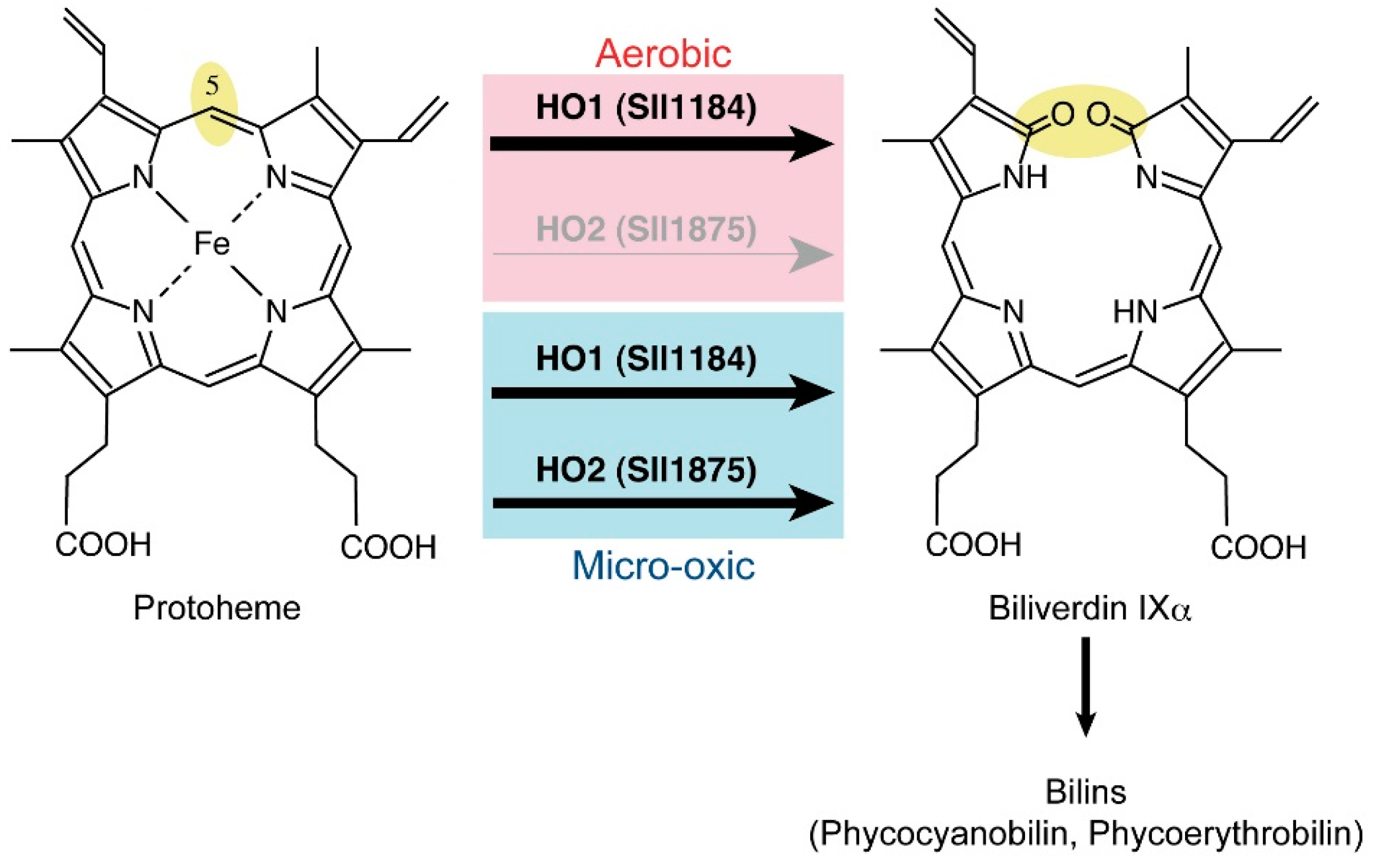
3. Transcriptional Regulator ChlR
3.1. Discovery of the ChlR Regulator
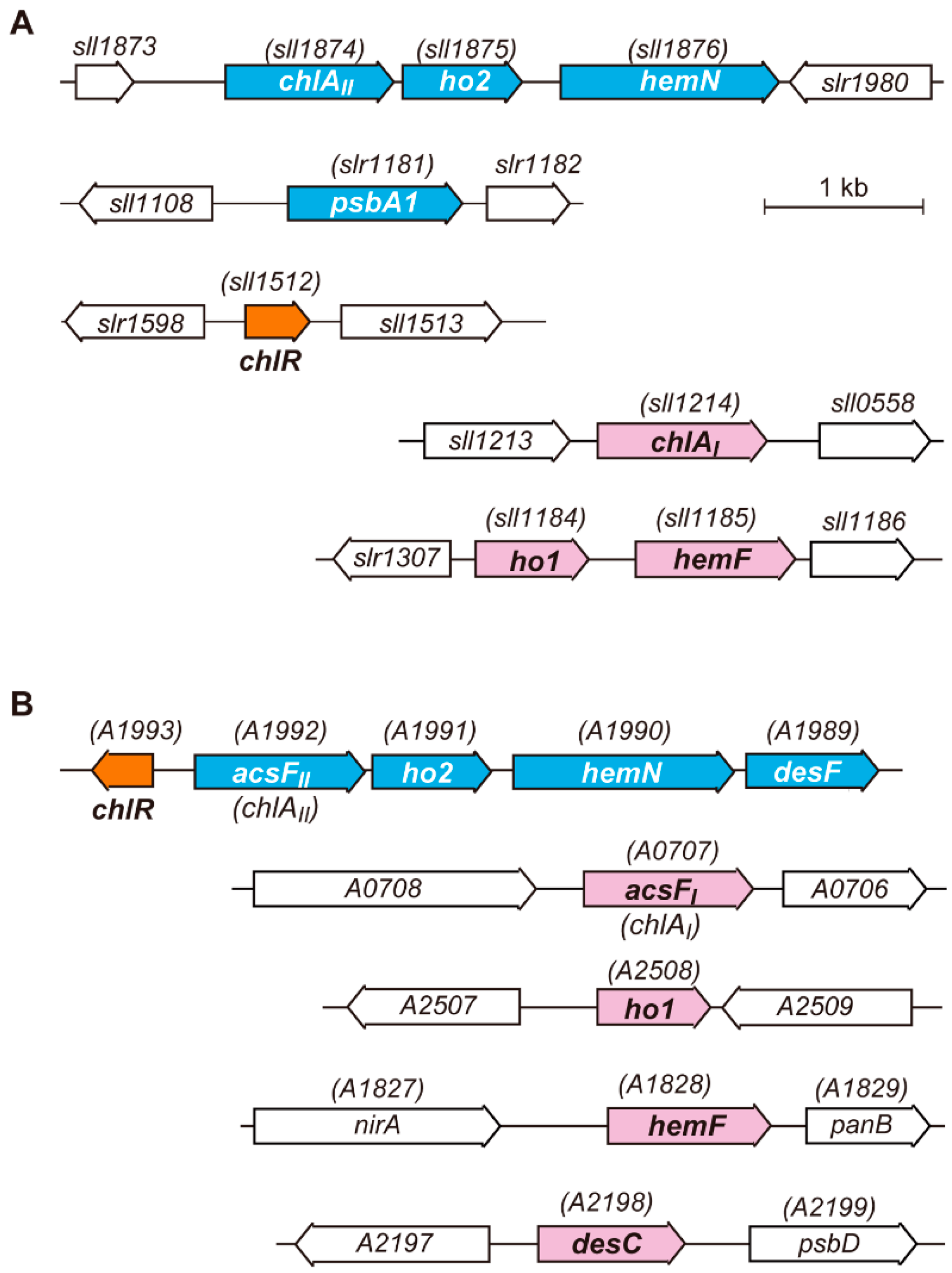
3.2. Characterization of ChlR

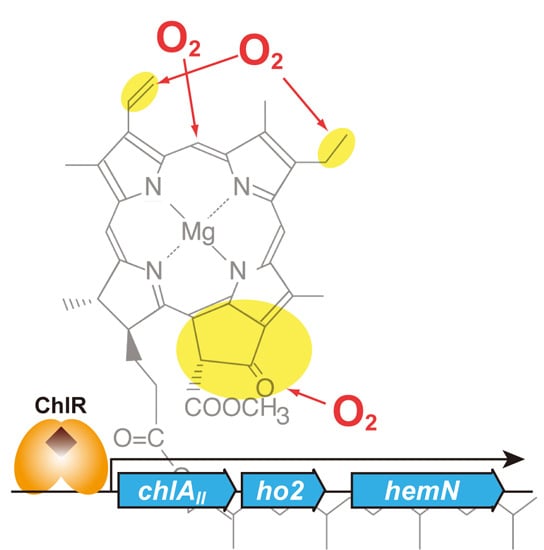
3.3. Target Genes of ChlR
4. Chlorophyll Biosynthesis under Nitrogen Fixing Conditions
5. Distribution of ChlR and Aerobic and Anaerobic Enzymes
6. GOE, Oxygen Crisis and Adaptive Evolution of Tetrapyrrole Biosynthesis
6.1. GOE and Oxygen Crisis
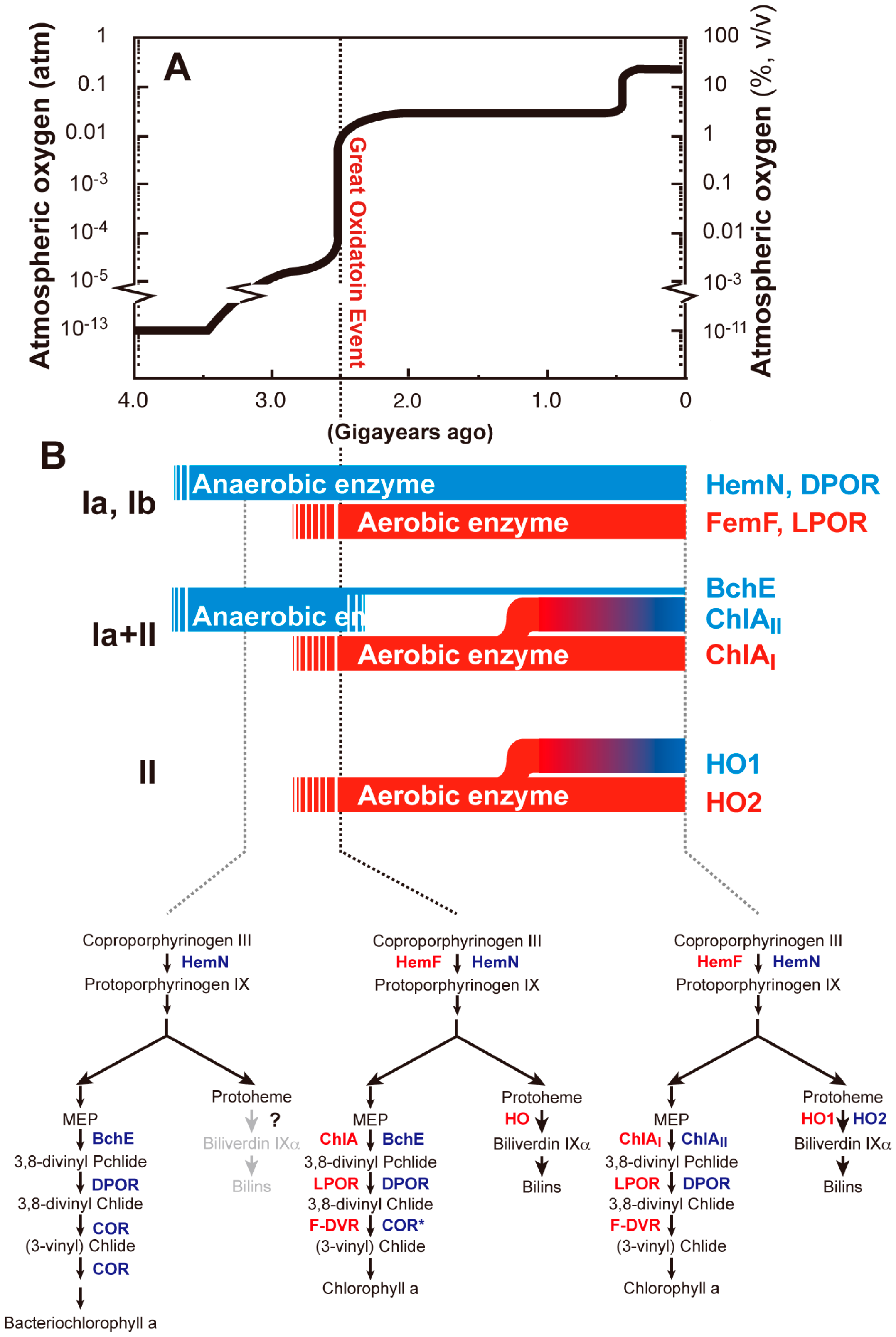
6.2. Adaptive Evolution of Tetrapyrrole Biosynthesis
| Class a | Reaction | Anaerobic Enzyme b | Aerobic Enzyme c | Extant Cyanobacteria d | Modes of Adaptive Evolution e | Regulator f | Ref. |
|---|---|---|---|---|---|---|---|
| Ia | CPgen oxidation | HemN | HemF * | HemN/HemF | Analogous, coexist | ChlR | [22,98] |
| Ib | Pchlide reduction | DPOR | LPOR g | DPOR/LPOR | Analogous, coexist h | PedR | [65,74,75] |
| 8-vinyl reduction | COR | N-DVR F-DVR i | F-DVR (N-DVR) j | Analogous, substitution | Unknown | - | |
| Ia + II | MPE cyclization | BchE k | ChlA * | ChlAII/ChlAI l | Analogous, substitution l; isoforms m | ChlR | [33,98] |
| II | Heme cleavage | Unknown | HO * | HO2/HO1 | Isoforms n | ChlR | [89,98] |
7. Perspectives and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gray, M.W.; Archibald, J.M. Origins of mitchondria and plastids. In Genomics of Chloroplasts and Mitochondria; Bock, R., Knoop, V., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 1–30. [Google Scholar]
- Stal, L.J.; Zehr, J.P. Cyanobacterial nitrogen fixation in the ocean: Diversity, regulation, and ecology. In The Cyanobacteria: Molecular Biology, Genomics and Evolution; Herrero, A., Flores, E., Eds.; Caister Academic Press: Norfolk, UK, 2008; pp. 423–446. [Google Scholar]
- Jørgensen, B.B.; Revsbech, N.P.; Blackburn, T.H.; Cohen, Y. Diurnal cycle of oxygen and sulfide microgradients and microbial photosynthesis in a cyanobacterial mat sediment. Appl. Environ. Microbiol. 1979, 38, 46–58. [Google Scholar]
- Steunou, A.S.; Bhaya, D.; Bateson, M.M.; Melendrez, M.C.; Ward, D.M.; Brecht, E.; Peters, J.W.; Kühl, M.; Grossman, A.R. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. USA 2006, 103, 2398–2403. [Google Scholar]
- Raymond, J.; Segrè, D. The effect of oxygen on biochemical networks and the evolution of complex life. Science 2006, 311, 1764–1767. [Google Scholar]
- Bauer, C.E.; Setterdahl, A.; Wu, J.; Robinson, B.R. Regulation of gene expression in response to oxygen tension. In The Purple Photosynthetic Bacteria; Hunter, C.N., Daldal, F., Thurnauer, M.C., Beatty, J.T., Eds.; Springer: Houten, The Netherlands, 2009; pp. 707–725. [Google Scholar]
- Bothe, H.; Schmitz, O.; Yates, M.G.; Newton, W.E. Nitrogen fixation and hydrogen metabolism in cyanobacteria. Microbiol. Mol. Biol. Rev. 2010, 74, 529–551. [Google Scholar]
- Tanaka, R.; Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007, 58, 321–346. [Google Scholar]
- Chew, A.G.; Bryant, D.A. Chlorophyll biosynthesis in bacteria: The origins of structural and functional diversity. Annu. Rev. Microbiol. 2007, 61, 113–129. [Google Scholar]
- Masuda, T.; Fujita, Y. Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 2008, 7, 1131–1149. [Google Scholar]
- Dailey, H. Terminal steps of haem biosynthesis. Biochem. Soc. Trans. 2002, 30, 590–595. [Google Scholar]
- Akhtar, M. Coproporphyrinogen III and protoporphyrinogen IX oxidases. In The Porphyrin Handbook, Volume 12/Iron and Cobalt Pigments: Biosynthesis, Structure, and Degradation; Kadish, K.M., Smith, K.M., Guilard, R., Eds.; Elsevier: New York, NY, USA, 2003; pp. 75–92. [Google Scholar]
- Cornah, J.E.; Smith, A.G. Transformation of uroporphyrinogen III into Protoheam. In Tetrapyrroles: Birth, Life and Death; Warren, M.J., Smith, A.G., Eds.; Springer: Austin, TX, USA, 2009; pp. 74–88. [Google Scholar]
- Troup, B.; Jahn, M.; Hungerer, C.; Jahn, D. Isolation of the hemF operon containing the gene for the Escherichia coli aerobic coproporphyrinogen III oxidase by in vivo complementation of a yeast hem13 mutant. J. Bacteriol. 1994, 176, 673–680. [Google Scholar]
- Phillips, J.D.; Whitby, F.G.; Warby, C.A.; Labbe, P.; Yang, C.; Pflugrath, J.W.; Ferrara, J.D.; Robinson, H.; Kushner, J.P.; Hill, C.P. Crystal structure of the oxygen-dependant coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 38960–38968. [Google Scholar]
- Dowling, D.P.; Vey, J.L.; Croft, A.K.; Drennan, C.L. Structural diversity in the AdoMet radical enzyme superfamily. Biochim. Biophys. Acta 2012, 1824, 1178–1195. [Google Scholar]
- Broderick, J.B.; Duffus, B.R.; Duschene, K.S.; Shepard, E.M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014, 114, 4229–4317. [Google Scholar]
- Troup, B.; Hungerer, C.; Jahn, D. Cloning and characterization of the Escherichia coli hemN gene encoding the oxygen-independent coproporphyrinogen III oxidase. J. Bacteriol. 1995, 177, 3326–3331. [Google Scholar]
- Layer, G.; Verfürth, K.; Mahlitz, E.; Jahn, D. Oxygen-independent coproporphyrinogen-III oxidase HemN from Escherichia coli. J. Biol. Chem. 2002, 277, 34136–34142. [Google Scholar]
- Layer, G.; Moser, J.; Heinz, D.W.; Jahn, D.; Schubert, W.D. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical sam enzymes. EMBO J. 2003, 22, 6214–6224. [Google Scholar]
- Layer, G.; Grage, K.; Teschner, T.; Schünemann, V.; Breckau, D.; Masoumi, A.; Jahn, M.; Heathcote, P.; Trautwein, A.; Jahn, D. Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: Functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-l-methionines. J. Biol. Chem. 2005, 280, 29038–29046. [Google Scholar]
- Goto, T.; Aoki, R.; Minamizaki, K.; Fujita, Y. Functional differentiation of two analogous coproporphyrinogen III oxidases for heme and chlorophyll biosynthesis pathways in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2010, 51, 650–663. [Google Scholar]
- Abicht, H.K.; Martinez, J.; Layer, G.; Jahn, D.; Solioz, M. Lactococcus lactis HemW (HemN) is a haem-binding protein with a putative role in haem trafficking. Biochem. J. 2012, 442, 335–343. [Google Scholar]
- Sasarman, A.; Letowski, J.; Czaika, G.; Ramirez, V.; Nead, M.A.; Jacobs, J.M.; Morais, R. Nucleotide sequence of the hemG gene involved in the protoporphyrinogen oxidase activity of Escherichia coli K12. Can. J. Microbiol. 1993, 39, 1155–1161. [Google Scholar]
- Hansson, M.; Hederstedt, L. Cloning and characterization of the Bacillus subtilis hemEHY gene cluster, which encodes protoheme IX biosynthetic enzymes. J. Bacteriol. 1992, 174, 8081–8093. [Google Scholar]
- Kato, K.; Tanaka, R.; Sano, S.; Tanaka, A.; Hosaka, H. Identification of a gene essential for protoporphyrinogen IX oxidase activity in the cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 2010, 107, 16649–16654. [Google Scholar]
- Koch, M.; Breithaupt, C.; Kiefersauer, R.; Freigang, J.; Huber, R.; Messerschmidt, A. Crystal structure of protoporphyrinogen IX oxidase: A key enzyme in haem and chlorophyll biosynthesis. EMBO J. 2004, 23, 1720–1728. [Google Scholar]
- Boynton, T.O.; Daugherty, L.E.; Dailey, T.A.; Dailey, H.A. Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry 2009, 48, 6705–6711. [Google Scholar]
- Kobayashi, K.; Masuda, T.; Tajima, N.; Wada, H.; Sato, N. Molecular phylogeny and intricate evolutionary history of the three isofunctional enzymes involved in the oxidation of protoporphyrinogen IX. Genome Biol. Evol. 2014, 6, 2141–2155. [Google Scholar]
- Bollivar, D.W. Intermediate steps in chlorophyll biosynthesis: Methylation and cyclization. In The Porphyrin Handbook, vol. 13, Chlorophylls and Bilins: Synthesis, and Degradation; Kadish, K.M., Smith, K.M., Guilard, R., Eds.; Elsevier: Amsterdam, The Netherlands, 2003; Volume 13, pp. 49–69. [Google Scholar]
- Bollivar, D.W.; Beale, S.I. The chlorophyll biosynthetic enzyme Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase (characterization and partial purification from Chlamydomonas reinhardtii and Synechocystis sp. PCC 6803). Plant Physiol. 1996, 112, 105–114. [Google Scholar]
- Pinta, V.; Picaud, M.; Reiss-Husson, F.; Astier, C. Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J. Bacteriol. 2002, 184, 746–753. [Google Scholar]
- Minamizaki, K.; Mizoguchi, T.; Goto, T.; Tamiaki, H.; Fujita, Y. Identification of two homologous genes, chlAI and chlAII, that are differentially involved in isocyclic ring formation of chlorophyll a in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 2008, 283, 2684–2692. [Google Scholar]
- Peter, E.; Salinas, A.; Wallner, T.; Jeske, D.; Dienst, D.; Wilde, A.; Grimm, B. Differential requirement of two homologous proteins encoded by sll1214 and sll1874 for the reaction of Mg protoporphyrin monomethylester oxidative cyclase under aerobic and micro-oxic growth conditions. Biochim. Biophys. Acta 2009, 1787, 1458–1467. [Google Scholar]
- Tottey, S.; Block, M.A.; Allen, M.; Westergren, T.; Albrieux, C.; Scheller, H.V.; Merchant, S.; Jensen, P.E. Arabidopsis Chl27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 2003, 100, 16119–16124. [Google Scholar]
- Moseley, J.; Quinn, J.; Eriksson, M.; Merchant, S. The crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 2000, 19, 2139–2151. [Google Scholar]
- Bollivar, D.W.; Braumann, I.; Berendt, K.; Gough, S.P.; Hansson, M. The Ycf54 protein is part of the membrane component of Mg-protoporphyrin IX monomethyl ester cyclase from barley (Hordeum vulgare L.). FEBS J. 2014, 281, 2377–2386. [Google Scholar]
- Gough, S.P.; Petersen, B.O.; Duus, J.O. Anaerobic chlorophyll isocyclic ring formation in Rhodobacter capsulatus requires a cobalamin cofactor. Proc. Natl. Acad. Sci. USA 2000, 97, 6908–6913. [Google Scholar]
- Nasrulhaq-Boyce, A.; Griffiths, W.T.; Jones, O.T. The use of continuous assays to characterize the oxidative cyclase that synthesizes the chlorophyll isocyclic ring. Biochem. J. 1987, 243, 23–29. [Google Scholar]
- Walker, C.J.; Mansfield, K.E.; Smith, K.M.; Castelfranco, P.A. Incorporation of atmospheric oxygen into the carbonyl functionality of the protochlorophyllide isocyclic ring. Biochem. J. 1989, 257, 599–602. [Google Scholar]
- Bollivar, D.W.; Beale, S.I. Formation of the isocyclic ring of chlorophyll by isolated Chlamydomonas reinhardtii chloroplasts. Photosynth. Res. 1995, 43, 113–124. [Google Scholar]
- Stenbaek, A.; Hansson, A.; Wulff, R.P.; Hansson, M.; Dietz, K.J.; Jensen, P.E. NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett. 2008, 582, 2773–2778. [Google Scholar]
- Hollingshead, S.; Kopecná, J.; Jackson, P.J.; Canniffe, D.P.; Davison, P.A.; Dickman, M.J.; Sobotka, R.; Hunter, C.N. Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J. Biol. Chem. 2012, 287, 27823–27833. [Google Scholar]
- Albus, C.A.; Salinas, A.; Czarnecki, O.; Kahlau, S.; Rothbart, M.; Thiele, W.; Lein, W.; Bock, R.; Grimm, B.; Schöttler, M.A. LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. 2012, 160, 1923–1939. [Google Scholar]
- Bollivar, D.; Suzuki, J.; Beatty, J.; Dobrowolski, J.; Bauer, C. Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J. Mol. Biol. 1994, 237, 622–640. [Google Scholar]
- Ouchane, S.; Steunou, A.; Picaud, M.; Astier, C. Aerobic and anaerobic Mg-protoporphyrin monomethyl ester cyclases in purple bacteria: A strategy adopted to bypass the repressive oxygen control system. J. Biol. Chem. 2004, 279, 6385–6394. [Google Scholar]
- Ouchane, S.; Picaud, M.; Therizols, P.; Reiss-Husson, F.; Astier, C. Global regulation of photosynthesis and respiration by FnrL: The first two targets in the tetrapyrrole pathway. J. Biol. Chem. 2007, 282, 7690–7699. [Google Scholar]
- Hassani, B.K.; Steunou, A.S.; Liotenberg, S.; Reiss-Husson, F.; Astier, C.; Ouchane, S. Adaptation to oxygen: Role of terminal oxidases in photosynthesis initiation in the purple photosynthetic bacterium, Rubrivivax gelatinosus. J. Biol. Chem. 2010, 285, 19891–19899. [Google Scholar]
- Tang, K.H.; Barry, K.; Chertkov, O.; Dalin, E.; Han, C.S.; Hauser, L.J.; Honchak, B.M.; Karbach, L.E.; Land, M.L.; Lapidus, A.; et al. Complete genome sequence of the filamentous anoxygenic phototrophic bacterium Chloroflexus aurantiacus. BMC Genomics 2011, 12. [Google Scholar] [CrossRef]
- Boldareva-Nuianzina, E.N.; Bláhová, Z.; Sobotka, R.; Koblízek, M. Distribution and origin of oxygen-dependent and oxygen-independent forms of Mg-protoporphyrin monomethylester cyclase among phototrophic proteobacteria. Appl. Environ. Microbiol. 2013, 79, 2596–2604. [Google Scholar]
- Masuda, T.; Takamiya, K. Novel insights into the enzymology, regulation and physiological functions of light-dependent protochlorophyllide oxidoreductase in angiosperms. Photosynth. Res. 2004, 81, 1–29. [Google Scholar]
- Heyes, D.; Hunter, C. Making light work of enzyme catalysis: Protochlorophyllide oxidoreductase. Trends Biochem. Sci. 2005, 30, 642–649. [Google Scholar]
- Fujita, Y. Protochlorophyllide reduction: A key step in the greening of plants. Plant Cell Physiol. 1996, 37, 411–421. [Google Scholar]
- Fujita, Y.; Bauer, C.E. The light-independent protochlorophyllide reductase: A nitrogenase-like enzyme catalyzing a key reaction for greening in the dark. In Chlorophylls and Bilins: Biosynthesis, Synthesis, and Degradation; Kadish, K., Smith, K.M., Guilard, R., Eds.; Academic Press: Amsterdom, The Netherlands, 2003; pp. 109–156. [Google Scholar]
- Reinbothe, C.; El Bakkouri, M.; Buhr, F.; Muraki, N.; Nomata, J.; Kurisu, G.; Fujita, Y.; Reinbothe, S. Chlorophyll biosynthesis: Spotlight on protochlorophyllide reduction. Trends Plant Sci. 2010, 15, 614–622. [Google Scholar]
- Fujita, Y.; Bauer, C. Reconstitution of light-independent protochlorophyllide reductase from purified BchL and BchN-BchB subunits. In vitro confirmation of nitrogenase-like features of a bacteriochlorophyll biosynthesis enzyme. J. Biol. Chem. 2000, 275, 23583–23588. [Google Scholar]
- Sarma, R.; Barney, B.; Hamilton, T.; Jones, A.; Seefeldt, L.; Peters, J. Crystal structure of the L protein of Rhodobacter sphaeroides light-independent protochlorophyllide reductase with MgADP bound: A homologue of the nitrogenase Fe protein. Biochemistry 2008, 47, 13004–13015. [Google Scholar]
- Muraki, N.; Nomata, J.; Ebata, K.; Mizoguchi, T.; Shiba, T.; Tamiaki, H.; Kurisu, G.; Fujita, Y. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 2010, 465, 110–114. [Google Scholar]
- Moser, J.; Lange, C.; Krausze, J.; Rebelein, J.; Schubert, W.D.; Ribbe, M.W.; Heinz, D.W.; Jahn, D. Structure of ADP-aluminium fluoride-stabilized protochlorophyllide oxidoreductase complex. Proc. Natl. Acad. Sci. USA 2013, 110, 2094–2098. [Google Scholar]
- Nomata, J.; Kitashima, M.; Inoue, K.; Fujita, Y. Nitrogenase Fe protein-like Fe-S cluster is conserved in L-protein (BchL) of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. FEBS Lett. 2006, 580, 6151–6154. [Google Scholar]
- Bröcker, M.; Virus, S.; Ganskow, S.; Heathcote, P.; Heinz, D.; Schubert, W.; Jahn, D.; Moser, J. ATP-driven reduction by dark-operative protochlorophyllide oxidoreductase from Chlorobium tepidum mechanistically resembles nitrogenase catalysis. J. Biol. Chem. 2008, 283, 10559–10567. [Google Scholar]
- Nomata, J.; Ogawa, T.; Kitashima, M.; Inoue, K.; Fujita, Y. NB-protein (BchN-BchB) of dark-operative protochlorophyllide reductase is the catalytic component containing oxygen-tolerant Fe-S clusters. FEBS Lett. 2008, 582, 1346–1350. [Google Scholar]
- Bröcker, M.; Wätzlich, D.; Uliczka, F.; Virus, S.; Saggu, M.; Lendzian, F.; Scheer, H.; Rüdiger, W.; Moser, J.; Jahn, D. Substrate recognition of nitrogenase-like dark operative protochlorophyllide oxidoreductase from Prochlorococcus marinus. J. Biol. Chem. 2008, 283, 29873–29881. [Google Scholar]
- Nomata, J.; Kondo, T.; Mizoguchi, T.; Tamiaki, H.; Itoh, S.; Fujita, Y. Dark-operative protochlorophyllide oxidoreductase generates substrate radicals by an iron-sulphur cluster in bacteriochlorophyll biosynthesis. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef]
- Yamazaki, S.; Nomata, J.; Fujita, Y. Differential operation of dual protochlorophyllide reductases for chlorophyll biosynthesis in response to environmental oxygen levels in the cyanobacterium Leptolyngbya boryana. Plant Physiol. 2006, 142, 911–922. [Google Scholar]
- Yamamoto, H.; Kurumiya, S.; Ohashi, R.; Fujita, Y. Oxygen sensitivity of a nitrogenase-like protochlorophyllide reductase from the cyanobacterium Leptolyngbya boryana. Plant Cell Physiol. 2009, 50, 1663–1673. [Google Scholar]
- Baker, M.E. Protochlorophyllide reductase is homologous to human carbonyl reductase and pig 20 β-hydroxysteroid dehydrogenase. Biochem. J. 1994, 300, 605–607. [Google Scholar]
- Labesse, G.; Vidal-Cros, A.; Chomilier, J.; Gaudry, M.; Mornon, J.P. Structural comparisons lead to the definition of a new superfamily of NAD(P)(H)-accepting oxidoreductases: The single-domain reductases/epimerases/dehydrogenases (the “RED” family). Biochem. J. 1994, 304, 95–99. [Google Scholar]
- Zhong, D. Ultrafast catalytic processes in enzymes. Curr. Opin. Chem. Biol. 2007, 11, 174–181. [Google Scholar]
- Wilks, H.; Timko, M. A light-dependent complementation system for analysis of NADPH: protochlorophyllide oxidoreductase: Identification and mutagenesis of two conserved residues that are essential for enzyme activity. Proc. Natl. Acad. Sci. USA 1995, 92, 724–728. [Google Scholar]
- Heyes, D.; Ruban, A.; Wilks, H.; Hunter, C. Enzymology below 200 K: The kinetics and thermodynamics of the photochemistry catalyzed by protochlorophyllide oxidoreductase. Proc. Natl. Acad. Sci. USA 2002, 99, 11145–11150. [Google Scholar]
- Heyes, D.; Heathcote, P.; Rigby, S.; Palacios, M.; van Grondelle, R.; Hunter, C. The first catalytic step of the light-driven enzyme protochlorophyllide oxidoreductase proceeds via a charge transfer complex. J. Biol. Chem. 2006, 281, 26847–26853. [Google Scholar]
- Sytina, O.; Heyes, D.; Hunter, C.; Alexandre, M.; van Stokkum, I.; van Grondelle, R.; Groot, M. Conformational changes in an ultrafast light-driven enzyme determine catalytic activity. Nature 2008, 456, 1001–1004. [Google Scholar]
- Fujita, Y.; Takagi, H.; Hase, T. Cloning of the gene encoding a protochlorophyllide reductase: The physiological significance of the co-existence of light-dependent and -independent protochlorophyllide reduction systems in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1998, 39, 177–185. [Google Scholar]
- Nakamura, K.; Hihara, Y. Photon flux density-dependent gene expression in Synechocystis sp. PCC 6803 is regulated by a small, redox-responsive, LuxR-type regulator. J. Biol. Chem. 2006, 281, 36758–36766. [Google Scholar]
- Horiuchi, M.; Nakamura, K.; Kojima, K.; Nishiyama, Y.; Hatakeyama, W.; Hisabori, T.; Hihara, Y. The PedR transcriptional regulator interacts with thioredoxin to connect photosynthesis with gene expression in cyanobacteria. Biochem. J. 2010, 431, 135–140. [Google Scholar]
- Shui, J.; Saunders, E.; Needleman, R.; Nappi, M.; Cooper, J.; Hall, L.; Kehoe, D.; Stowe-Evans, E. Light-dependent and light-independent protochlorophyllide oxidoreductases in the chromatically adapting cyanobacterium Fremyella diplosiphon UTEX 481. Plant Cell Physiol. 2009, 50, 1507–1521. [Google Scholar]
- Nagata, N.; Tanaka, R.; Satoh, S.; Tanaka, A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 2005, 17, 233–240. [Google Scholar]
- Chew, A.G.; Bryant, D.A. Characterization of a plant-like protochlorophyllide a divinyl reductase in green sulfur bacteria. J. Biol. Chem. 2007, 282, 2967–2975. [Google Scholar]
- Canniffe, D.P.; Jackson, P.J.; Hollingshead, S.; Dickman, M.J.; Hunter, C.N. Identification of an 8-vinyl reductase involved in bacteriochlorophyll biosynthesis in Rhodobacter sphaeroides and evidence for the existence of a third distinct class of the enzyme. Biochem. J. 2013, 450, 397–405. [Google Scholar]
- Islam, M.R.; Aikawa, S.; Midorikawa, T.; Kashino, Y.; Satoh, K.; Koike, H. Slr1923 of Synechocystis sp. PCC6803 is essential for conversion of 3,8-divinyl(proto)chlorophyll(ide) to 3-monovinyl(proto)chlorophyll(ide). Plant Physiol. 2008, 148, 1068–1081. [Google Scholar]
- Ito, H.; Yokono, M.; Tanaka, R.; Tanaka, A. Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC 6803. J. Biol. Chem. 2008, 283, 9002–9011. [Google Scholar]
- Ito, H.; Tanaka, A. Evolution of a new chlorophyll metabolic pathway driven by the dynamic changes in enzyme promiscuous activity. Plant Cell Physiol. 2014, 55, 593–603. [Google Scholar]
- Tsukatani, Y.; Yamamoto, H.; Harada, J.; Yoshitomi, T.; Nomata, J.; Kasahara, M.; Mizoguchi, T.; Fujita, Y.; Tamiaki, H. An unexpectedly branched biosynthetic pathway for bacteriochlorophyll b capable of absorbing near-infrared light. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef]
- Nomata, J.; Mizoguchi, T.; Tamiaki, H.; Fujita, Y. A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis—reconstitution of chlorophyllide a reductase with purified X-protein (BchX) and YZ-protein (BchY-BchZ) from Rhodobacter capsulatus. J. Biol. Chem. 2006, 281, 15021–15028. [Google Scholar]
- Yamamoto, H.; Kato, M.; Yamanashi, K.; Fujita, Y. Reconstitution of a sequential reaction of two nitrogenase-like enzymes in the bacteriochlorophyll biosynthetic pathway of Rhodobacter capsulatus. Biochem. Biophys. Res. Commun. 2014, 448, 200–205. [Google Scholar]
- Harada, J.; Mizoguchi, T.; Tsukatani, Y.; Yokono, M.; Tanaka, A.; Tamiaki, H. Chlorophyllide a oxidoreductase works as one of the divinyl reductases specifically involved in bacteriochlorophyll a biosynthesis. J. Biol. Chem. 2014, 289, 12716–12726. [Google Scholar]
- Kim, E.J.; Kim, J.S.; Rhee, H.J.; Lee, J.K. Growth arrest of Synechocystis sp. PCC 6803 by superoxide generated from heterologously expressed Rhodobacter sphaeroides chlorophyllide a reductase. FEBS Lett. 2009, 583, 219–223. [Google Scholar]
- Aoki, R.; Goto, T.; Fujita, Y. A heme oxygenase isoform is essential for aerobic growth in the cyanobacterium Synechocystis sp. PCC 6803: Modes of differential operation of two isoforms/enzymes to adapt to low oxygen environments in cyanobacteria. Plant Cell Physiol. 2011, 52, 1744–1756. [Google Scholar]
- Cornejo, J.; Willows, R.D.; Beale, S.I. Phytobilin biosynthesis: Cloning and expression of a gene encoding soluble ferredoxin-dependent heme oxygenase from Synechocystis sp. PCC 6803. Plant J. 1998, 15, 99–107. [Google Scholar]
- Willows, R.D.; Mayer, S.M.; Foulk, M.S.; DeLong, A.; Hanson, K.; Chory, J.; Beale, S.I. Phytobilin biosynthesis: The Synechocystis sp. PCC 6803 heme oxygenase-encoding ho1 gene complements a phytochrome-deficient Arabidopsis thalianna hy1 mutant. Plant Mol. Biol. 2000, 43, 113–120. [Google Scholar]
- Zhang, X.; Migita, C.T.; Sato, M.; Sasahara, M.; Yoshida, T. Protein expressed by the ho2 gene of the cyanobacterium Synechocystis sp. PCC 6803 is a true heme oxygenase. Properties of the heme and enzyme complex. FEBS J. 2005, 272, 1012–1022. [Google Scholar]
- Sugishima, M.; Hagiwara, Y.; Zhang, X.; Yoshida, T.; Migita, C.T.; Fukuyama, K. Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme. Biochemistry 2005, 44, 4257–4266. [Google Scholar]
- Sugishima, M.; Migita, C.T.; Zhang, X.; Yoshida, T.; Fukuyama, K. Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC 6803 in complex with heme. Eur. J. Biochem. 2004, 271, 4517–4525. [Google Scholar]
- Yilmaz, M.; Kang, I.; Beale, S.I. Heme oxygenase 2 of the cyanobacterium Synechocystis sp. PCC 6803 is induced under a microaerobic atmosphere and is required for microaerobic growth at high light intensity. Photosynth. Res. 2010, 103, 47–59. [Google Scholar]
- Brüggemann, H.; Bauer, R.; Raffestin, S.; Gottschalk, G. Characterization of a heme oxygenase of Clostridium tetani and its possible role in oxygen tolerance. Arch. Microbiol. 2004, 182, 259–263. [Google Scholar]
- Wegiel, B.; Nemeth, Z.; Correa-Costa, M.; Bulmer, A.C.; Otterbein, L.E. Heme oxygenase-1: A metabolic nike. Antioxid. Redox Signal 2014, 20, 1709–1722. [Google Scholar]
- Aoki, R.; Takeda, T.; Omata, T.; Ihara, K.; Fujita, Y. MarR-type transcriptional regulator ChlR activates expression of tetrapyrrole biosynthesis genes in response to low-oxygen conditions in cyanobacteria. J. Biol. Chem. 2012, 287, 13500–13507. [Google Scholar]
- Ludwig, M.; Pandelia, M.E.; Chew, C.Y.; Zhang, B.; Golbeck, J.H.; Krebs, C.; Bryant, D.A. ChlR protein of Synechococcus sp. PCC 7002 is a transcription activator that uses an oxygen-sensitive [4Fe-4S] cluster to control genes involved in pigment biosynthesis. J. Biol. Chem. 2014, 289, 16624–16639. [Google Scholar]
- Tsujimoto, R.; Kamiya, N.; Fujita, Y. Transcriptional regulators ChlR and CnfR are essential for diazotrophic growth in nonheterocystous cyanobacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 6762–6767. [Google Scholar]
- Ellison, D.W.; Miller, V.L. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 2006, 9, 153–159. [Google Scholar]
- Perera, I.C.; Grove, A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell Biol. 2010, 2, 243–254. [Google Scholar]
- Poor, C.B.; Chen, P.R.; Duguid, E.; Rice, P.A.; He, C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J. Biol. Chem. 2009, 284, 23517–23524. [Google Scholar]
- Crack, J.C.; Green, J.; Hutchings, M.I.; Thomson, A.J.; Le Brun, N.E. Bacterial iron-sulfur regulatory proteins as biological sensor-switches. Antioxid. Redox Signal 2012, 17, 1215–1231. [Google Scholar]
- Mohamed, A.; Jansson, C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 1989, 13, 693–700. [Google Scholar]
- Summerfield, T.C.; Toepel, J.; Sherman, L.A. Low-oxygen induction of normally cryptic psbA genes in cyanobacteria. Biochemistry 2008, 47, 12939–12941. [Google Scholar]
- Miller, S.R.; Bebout, B.M. Variation in sulfide tolerance of photosystem II in phylogenetically diverse cyanobacteria from sulfidic habitats. Appl. Environ. Microbiol. 2004, 70, 736–744. [Google Scholar]
- Aoki, R.; Hiraide, Y.; Yamakawa, H.; Fujita, Y. A novel “oxygen-induced” greening process in a cyanobacterial mutant lacking the transcriptional activator ChlR involved in low-oxygen adaptation of tetrapyrrole biosynthesis. J. Biol. Chem. 2014, 289, 1841–1851. [Google Scholar]
- Summerfield, T.C.; Nagarajan, S.; Sherman, L. Gene expression under low oxygen conditions in the cyanobacterium Synechocystis sp. PCC 6803 demonstrates Hik31-dependent and -independent responses. Microbiology 2012, 157, 301–312. [Google Scholar]
- Seefeldt, L.C.; Hoffman, B.M.; Dean, D.R. Electron transfer in nitrogenase catalysis. Curr. Opin. Chem. Biol. 2012, 16, 19–25. [Google Scholar]
- Eady, R.R.; Smith, B.E.; Cook, K.A.; Postgate, J.R. Nitrogenase of Klebsiella pneumoniae. Purification and properties of the component proteins. Biochem. J. 1972, 128, 655–675. [Google Scholar]
- Kumar, K.; Mella-Herrera, R.A.; Golden, J.W. Cyanobacterial heterocysts. Cold Spring Harb. Perspect Biol. 2010, 2. [Google Scholar] [CrossRef]
- Sugiura, K.; Itoh, S. Single-cell confocal spectrometry of a filamentous cyanobacterium Nostoc at room and cryogenic temperature. Diversity and differentiation of pigment systems in 311 cells. Plant Cell Physiol. 2012, 53, 1492–1506. [Google Scholar]
- Awai, K.; Wolk, C.P. Identification of the glycosyl transferase required for synthesis of the principal glycolipid characteristic of heterocysts of Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 2007, 266, 98–102. [Google Scholar]
- Valladares, A.; Herrero, A.; Pils, D.; Schmetterer, G.; Flores, E. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol. Microbiol. 2003, 47, 1239–1249. [Google Scholar]
- Weare, N.M.; Benemann, J.R. Nitrogenase activity and photosynthesis in Plectonema boryanum. J. Bacteriol. 1974, 119, 258–265. [Google Scholar]
- Falkowski, P.G.; Godfrey, L.V. Electrons, life and the evolution of Earth’s oxygen cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 2705–2716. [Google Scholar]
- Kasting, J.F.; Siefert, J.L. Life and the evolution of Earth’s atmosphere. Science 2002, 296, 1066–1068. [Google Scholar]
- Sessions, A.L.; Doughty, D.M.; Welander, P.V.; Summons, R.E.; Newman, D.K. The continuing puzzle of the great oxidation event. Curr. Biol. 2009, 19, R567–R574. [Google Scholar]
- Imlay, J.A. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar]
- Raymond, J.; Blankenship, R.E. Biosynthetic pathways, gene replacement and the antiquity of life. Geobiology 2004, 2, 199–203. [Google Scholar]
- Galperin, M.Y.; Walker, D.R.; Koonin, E.V. Analogous enzymes: Independent inventions in enzyme evolution. Genome Res. 1998, 8, 779–790. [Google Scholar]
- Kaschner, M.; Loeschcke, A.; Krause, J.; Minh, B.Q.; Heck, A.; Endres, S.; Svensson, V.; Wirtz, A.; von Haeseler, A.; Jaeger, K.E.; et al. Discovery of the first light-dependent protochlorophyllide oxidoreductase in anoxygenic phototrophic bacteria. Mol. Microbiol. 2014, 93, 1066–1078. [Google Scholar]
- Saunders, A.H.; Golbeck, J.H.; Bryant, D.A. Characterization of bcib: A ferredoxin-dependent 8-vinyl-protochlorophyllide reductase from the green sulfur bacterium Chloroherpeton thalassium. Biochemistry 2013, 52, 8442–8451. [Google Scholar]
- Fixen, K.R.; Baker, A.W.; Stojkovic, E.A.; Beatty, J.T.; Harwood, C.S. Apo-bacteriophytochromes modulate bacterial photosynthesis in response to low light. Proc. Natl. Acad. Sci. USA 2014, 111, E237–E244. [Google Scholar]
- Liu, X.; Sheng, J.; Curtiss, R. Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 6899–6904. [Google Scholar]
- Savage, N. Algae: The scum solution. Nature 2011, 474, S15–S16. [Google Scholar]
- Oliver, J.W.; Atsumi, S. Metabolic design for cyanobacterial chemical synthesis. Photosynth. Res. 2014, 120, 249–261. [Google Scholar]
- Sakurai, H.; Masukawa, H. Promoting R&D in photobiological hydrogen production utilizing mariculture-raised cyanobacteria. Mar. Biotechnol. 2007, 9, 128–145. [Google Scholar]
- Sakurai, H.; Masukawa, H.; Kitashima, M.; Inoue, K. A feasibility study of large-scale photobiological hydrogen production utilizing mariculture-raised cyanobacteria. Adv. Exp. Med. Biol. 2010, 675, 291–303. [Google Scholar]
- Masukawa, H.; Kitashima, M.; Inoue, K.; Sakurai, H.; Hausinger, R.P. Genetic engineering of cyanobacteria to enhance biohydrogen production from sunlight and water. AmBio 2012, 41, 169–173. [Google Scholar]
- Rögner, M. Metabolic engineering of cyanobacteria for the production of hydrogen from water. Biochem. Soc. Trans. 2013, 41, 1254–1259. [Google Scholar]
- Watabe, K.; Mimuro, M.; Tsuchiya, T. Development of a highly-frequent in vivo transposon mutagenesis system for Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942. Plant Cell Physiol. 2014, 55, 2017–2026. [Google Scholar]
- Hiraide, Y.; Oshima, K.; Fujisawa, T.; Uesaka, K.; Hirose, Y.; Tsujimoto, R.; Yamamoto, H.; Okamoto, S.; Nakamura, Y.; Terauchi, K.; et al. Loss of cytochrome cM stimulates cyanobacterial heterotrophic growth in the dark. Plant Cell Physiol. 2015, 56, 334–345. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujita, Y.; Tsujimoto, R.; Aoki, R. Evolutionary Aspects and Regulation of Tetrapyrrole Biosynthesis in Cyanobacteria under Aerobic and Anaerobic Environments. Life 2015, 5, 1172-1203. https://doi.org/10.3390/life5021172
Fujita Y, Tsujimoto R, Aoki R. Evolutionary Aspects and Regulation of Tetrapyrrole Biosynthesis in Cyanobacteria under Aerobic and Anaerobic Environments. Life. 2015; 5(2):1172-1203. https://doi.org/10.3390/life5021172
Chicago/Turabian StyleFujita, Yuichi, Ryoma Tsujimoto, and Rina Aoki. 2015. "Evolutionary Aspects and Regulation of Tetrapyrrole Biosynthesis in Cyanobacteria under Aerobic and Anaerobic Environments" Life 5, no. 2: 1172-1203. https://doi.org/10.3390/life5021172
APA StyleFujita, Y., Tsujimoto, R., & Aoki, R. (2015). Evolutionary Aspects and Regulation of Tetrapyrrole Biosynthesis in Cyanobacteria under Aerobic and Anaerobic Environments. Life, 5(2), 1172-1203. https://doi.org/10.3390/life5021172