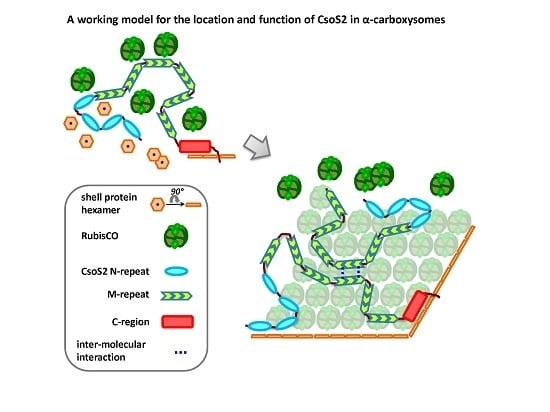
Advances in Understanding Carboxysome Assembly in Prochlorococcus and Synechococcus Implicate CsoS2 as a Critical Component
Abstract
:1. Introduction
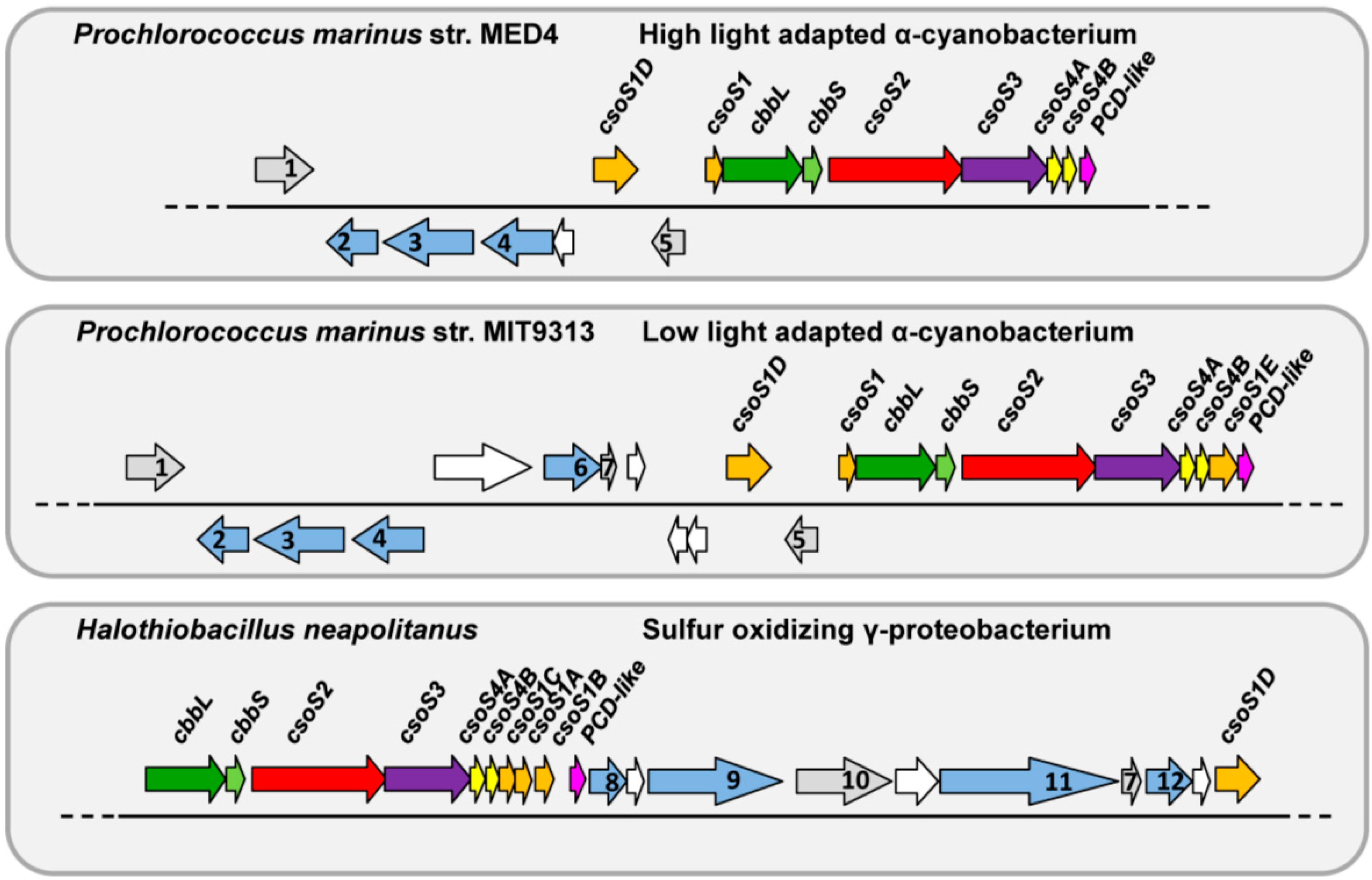
2. Results
2.1. The Hnea csoS2 Gene Inherently Encodes Two Protein Products
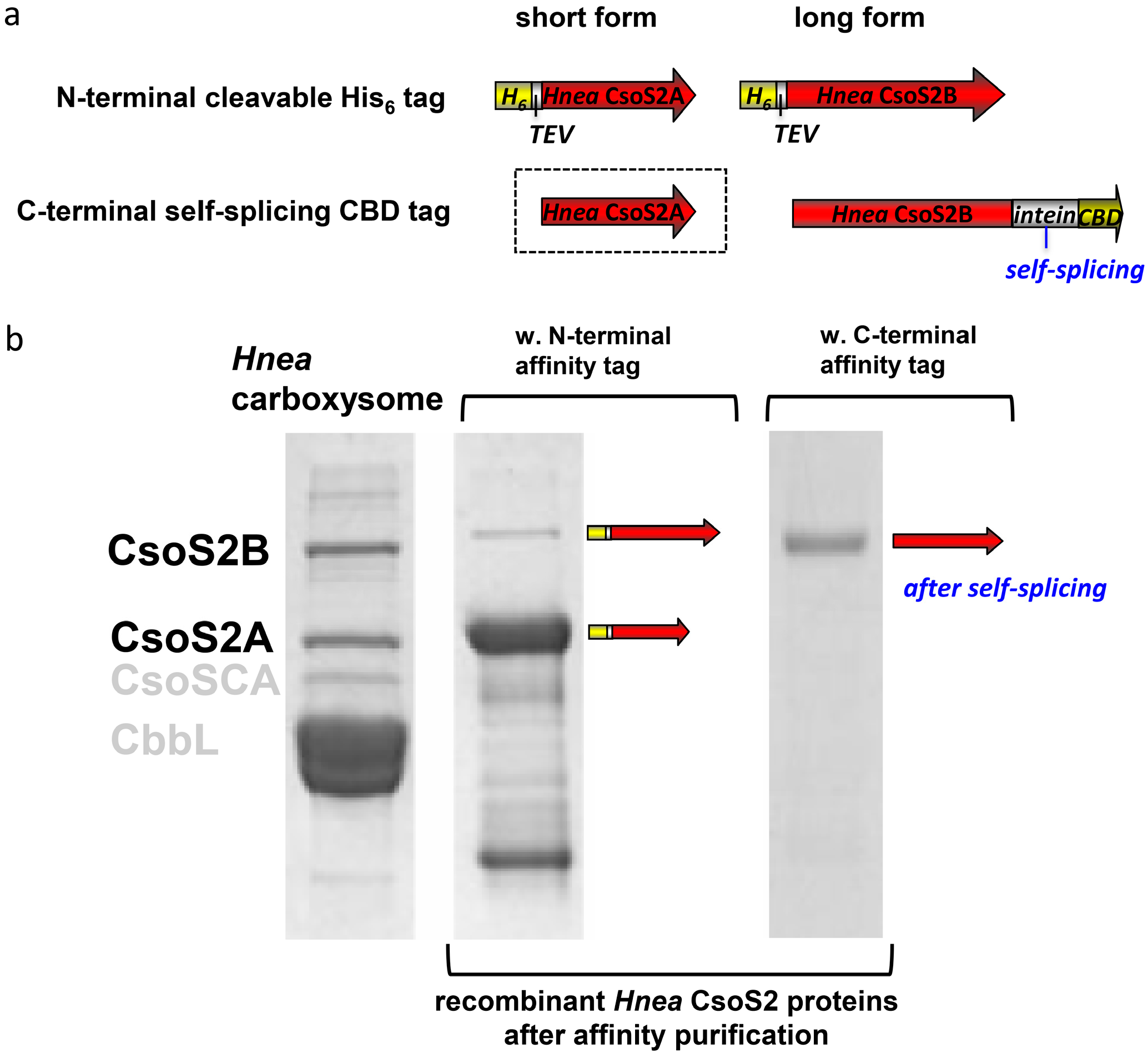
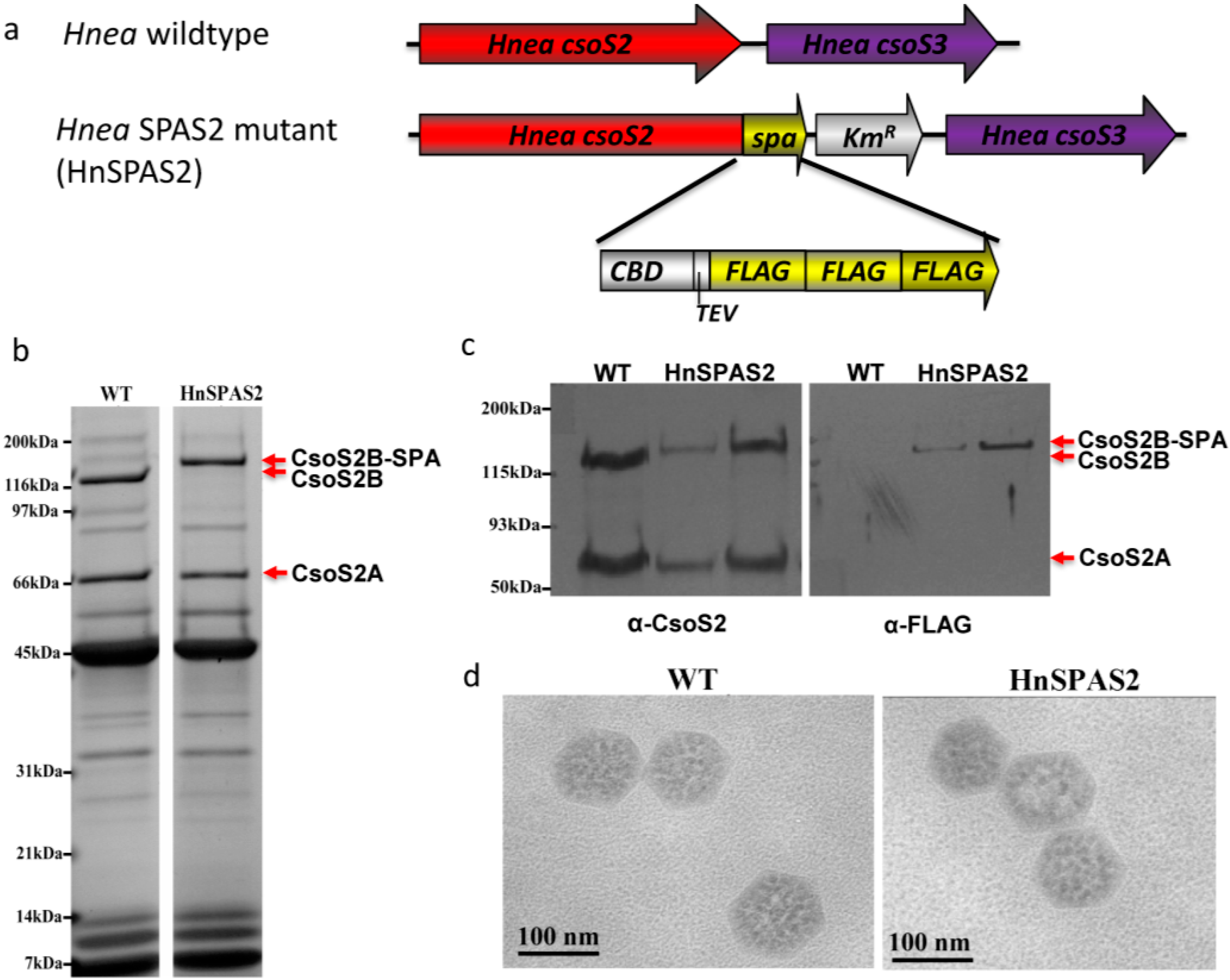
2.2. A CsoS2 Knockout Mutant Lack carboxysomes

2.3. Hints about the Location of CsoS2 Protein in Hnea carboxysomes
2.4. SPA-Tagged Hnea carboxysomes Can Be Selectively Purified by Affinity Chromatography
2.5. Unique Primary Structures of Prochlorococcus CsoS2s
| CsoS2 from | N-repeats | M-repeats * | C-region intact ? | length (aa) | calculated pI | Residues Count | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | H | R | K | E | Q | ||||||
| Prochlorococcus marinus str. MED4 | 4 | 6 (TTNTTT) | Y | 765 | 9.63 | 12 | 4 | 43 | 78 | 41 | 31 |
| Prochlorococcus marinus str. MIT9313 | 4 | 6 (TTNTTT) | Y | 792 | 9.75 | 12 | 6 | 67 | 44 | 45 | 48 |
| Halothiobacillus neapolitanus C2 | 4 | 6 (TTNTTT) | Y | 869 | 9.06 | 20 | 14 | 63 | 38 | 49 | 51 |
| Thioalkalivibrio sp. ALR17-21 | 5 | 8 (TTTTTTTT) | Y | 1014 | 7.08 | 22 | 22 | 106 | 26 | 87 | 44 |
| Thioalkalivibrio nitratireducens DSM14787 | 4 | 6 (TTNTTT) | Y | 782 | 9.19 | 15 | 4 | 78 | 34 | 46 | 34 |
| Bradyrhizobium sp. BTAi1 | 2 | 5 (SNSNN) | Y | 563 | 10.99 | 2 | 8 | 53 | 12 | 20 | 29 |
| Thermithiobacillus tepidarius DSM 3134 | 4 | 7 (TTNTNTS) | Y | 847 | 9.90 | 14 | 8 | 79 | 25 | 53 | 37 |
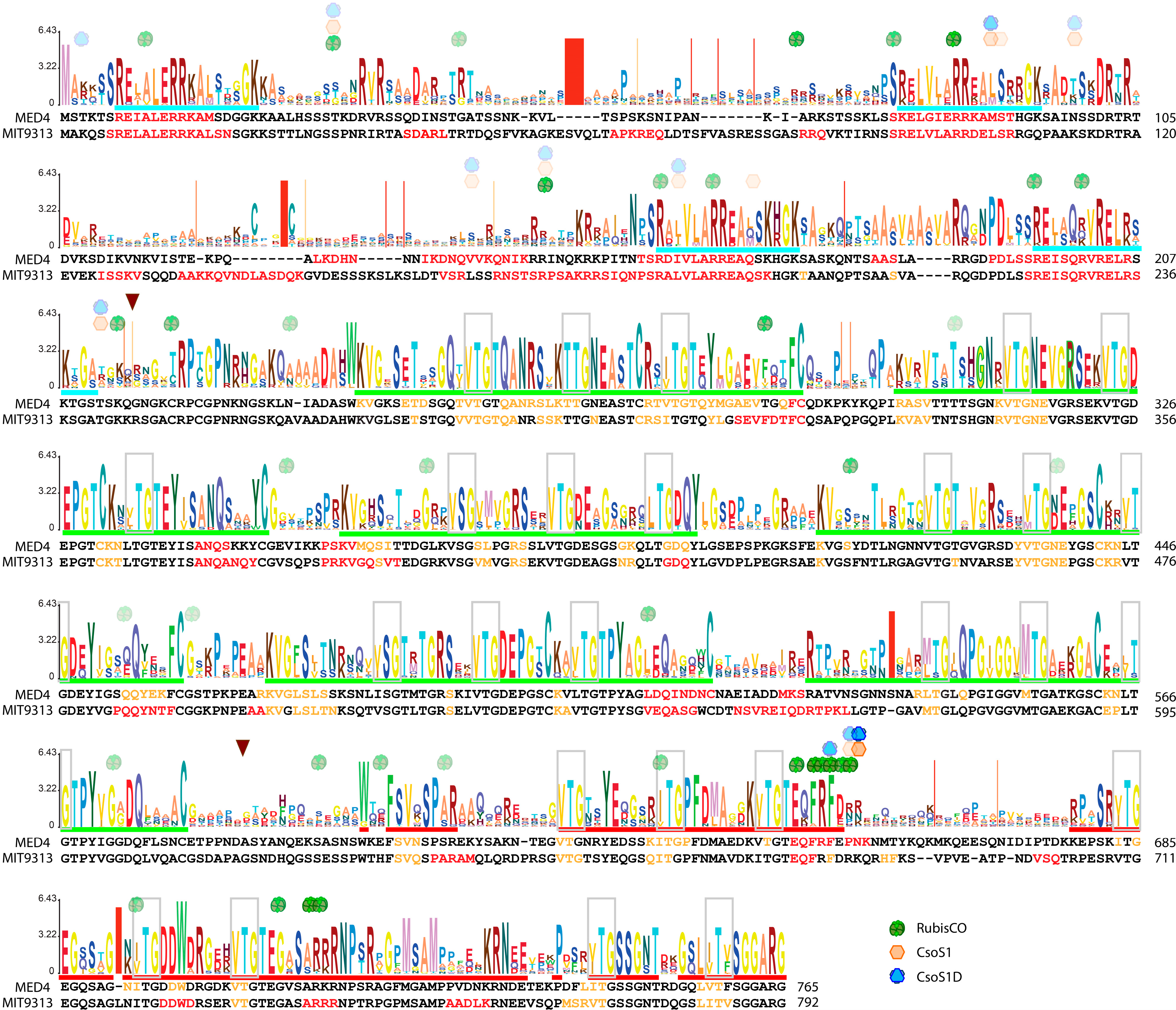
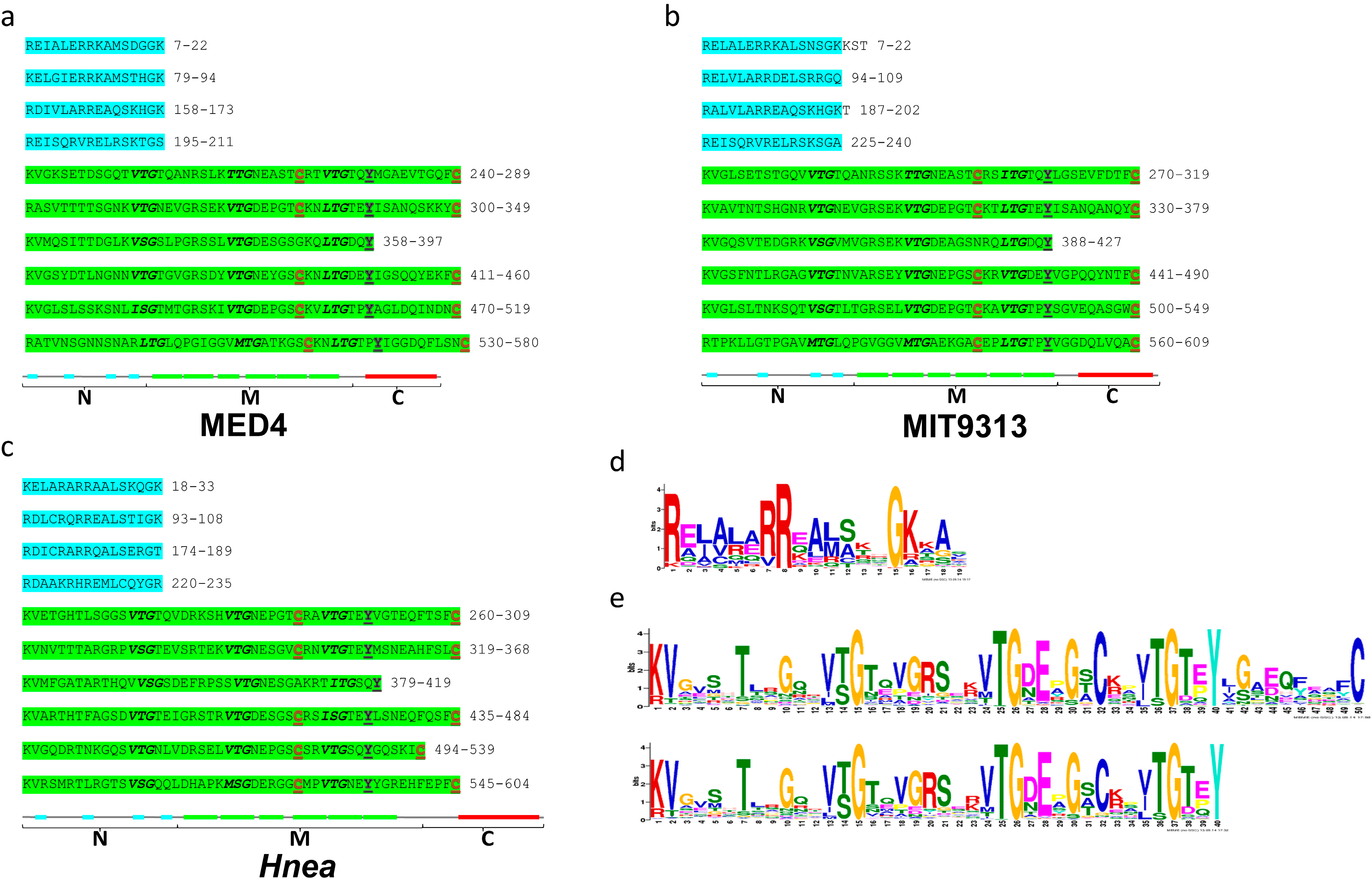
2.6. The Three-Region Architecture of CsoS2 Is Conserved
| MED4 | MIT9313 | Hnea | ALR17-21 | Tni | Brady | |
|---|---|---|---|---|---|---|
| C-region: I/S | ||||||
| MED4 | ||||||
| MIT9313 | 58.5/73.3 | |||||
| Hnea | 34.7/45.3 | 37.3/49.4 | ||||
| ALR17-21 | 38.1/49.7 | 43.2/54.7 | 42.5/56.9 | |||
| Tni | 51.7/60.7 | 58.2/68.8 | 40.7/49.4 | 44.0/56.0 | ||
| Brady | 34.3/47.1 | 34.8/45.9 | 26.9/37.5 | 30.8/41.8 | 37.6/45.4 | |
| Tte | 38.6/54.3 | 44.8/60.4 | 41.4/56.1 | 43.8/57.6 | 45.1/53.5 | 35.1/47.0 |
| full-length: I/S | ||||||
| MED4 | ||||||
| MIT9313 | 54.1/69.5 | |||||
| Hnea | 30.8/45.6 | 32.3/46.4 | ||||
| ALR17-21 | 29.2/42.0 | 29.2/40.3 | 33.1/47.3 | |||
| Tni | 41.2/56.4 | 45.0/59.5 | 29.7/39.5 | 32.1/43.1 | ||
| Brady | 22.8/32.8 | 22.1/31.4 | 20.6/30.6 | 19.8/27.5 | 26.2/35.6 | |
| Tte | 28.7/42.7 | 31.6/43.7 | 41.7/54.7 | 35.8/47.6 | 34.4/44.1 | 22.6/32.7 |
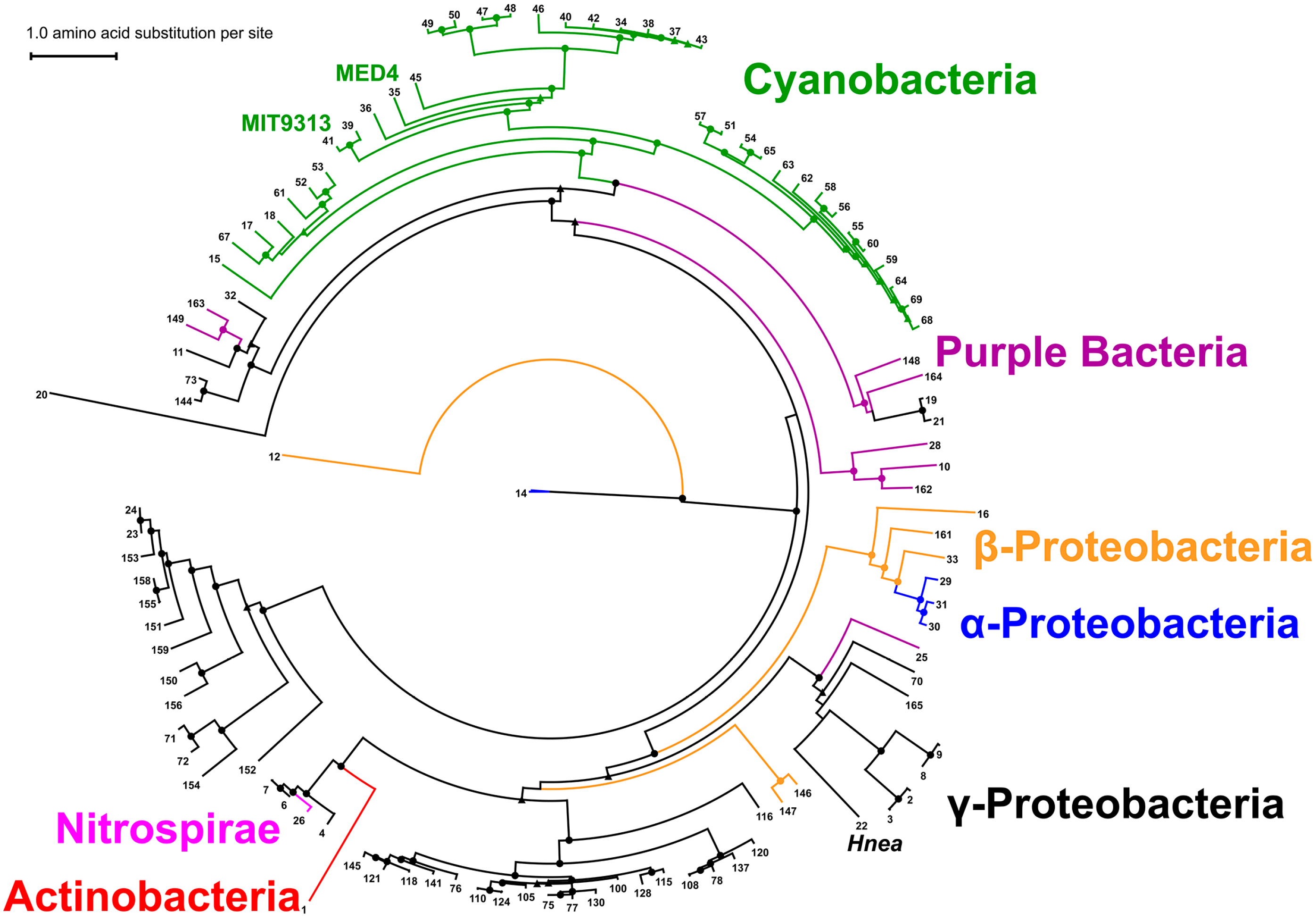
2.7. Phylogenetic Analysis of CsoS2 Hints at the Origin of α-carboxysomes
2.8. Structure Prediction of CsoS2: An Intrinsically Disordered Protein?

2.9. Experimental Data Supports the Prediction of a Highly Flexible CsoS2
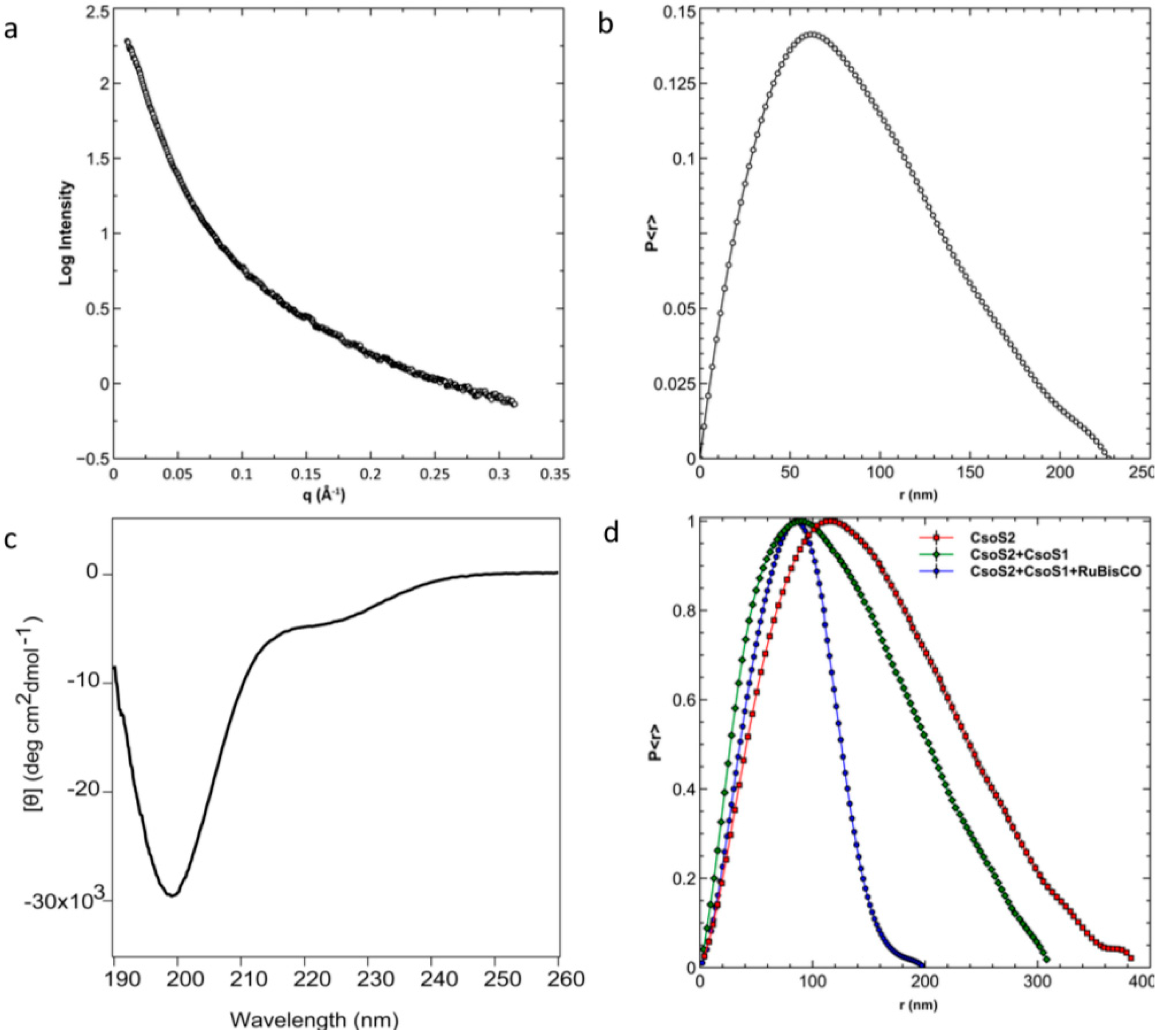
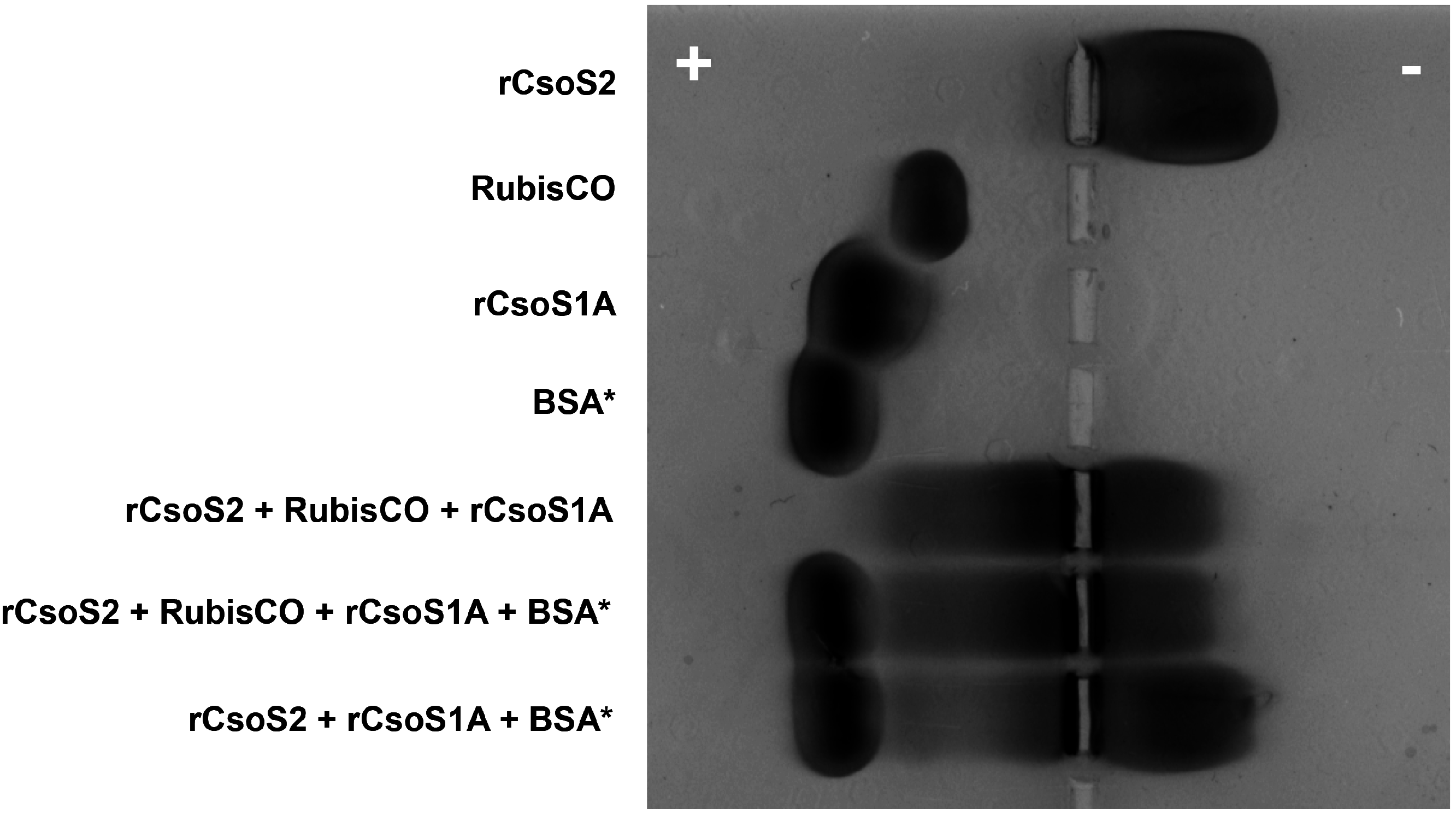
2.10. CsoS2 Interacts with the Major Shell Protein CsoS1 and RuBisCO in vitro
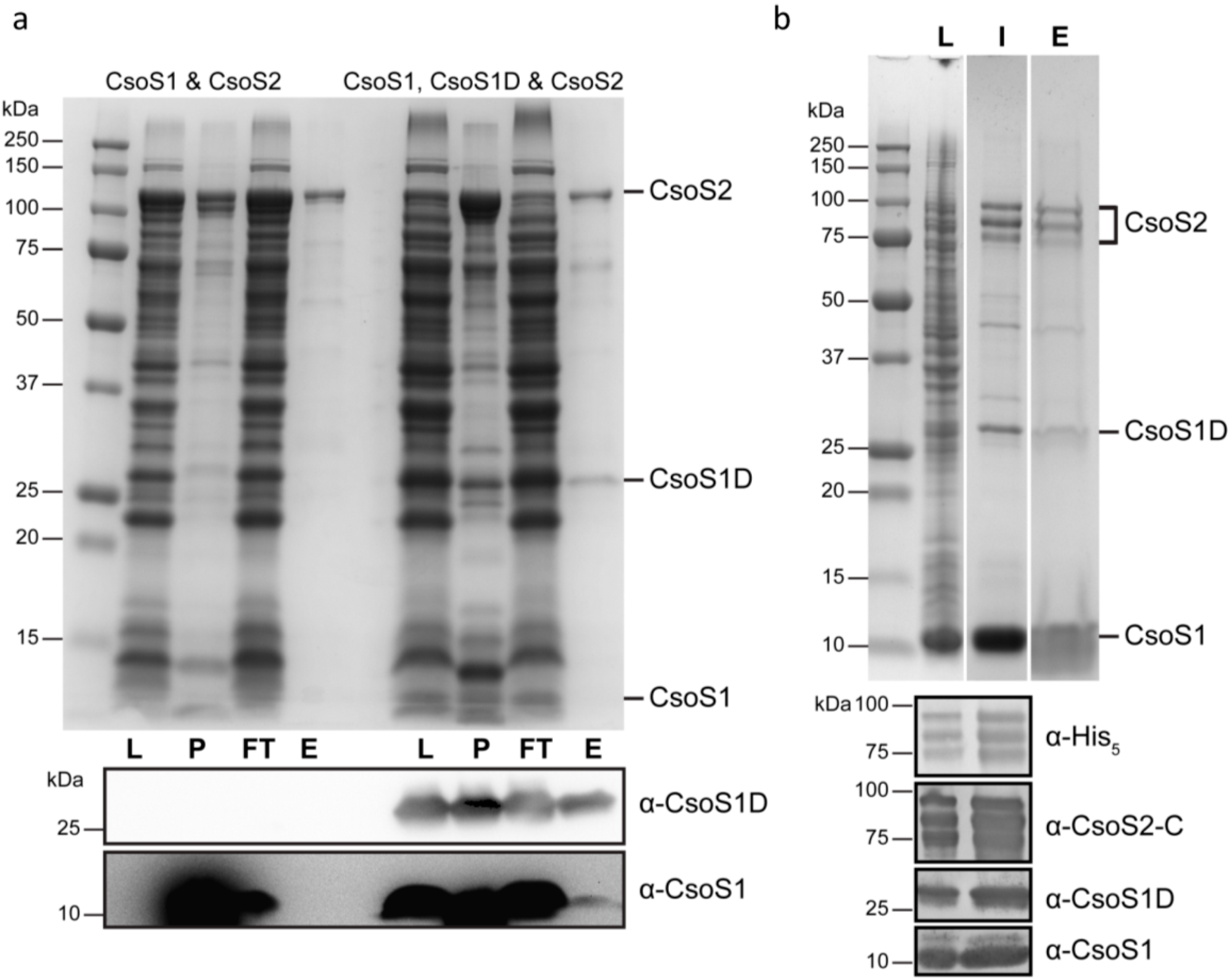
2.11. In vivo Formation of CsoS2-CsoS1 Protein Complexes in the Presence of CsoS1D in E. coli
2.12. Exploring the CsoS2 Protein Interaction Network Using Peptide Arrays
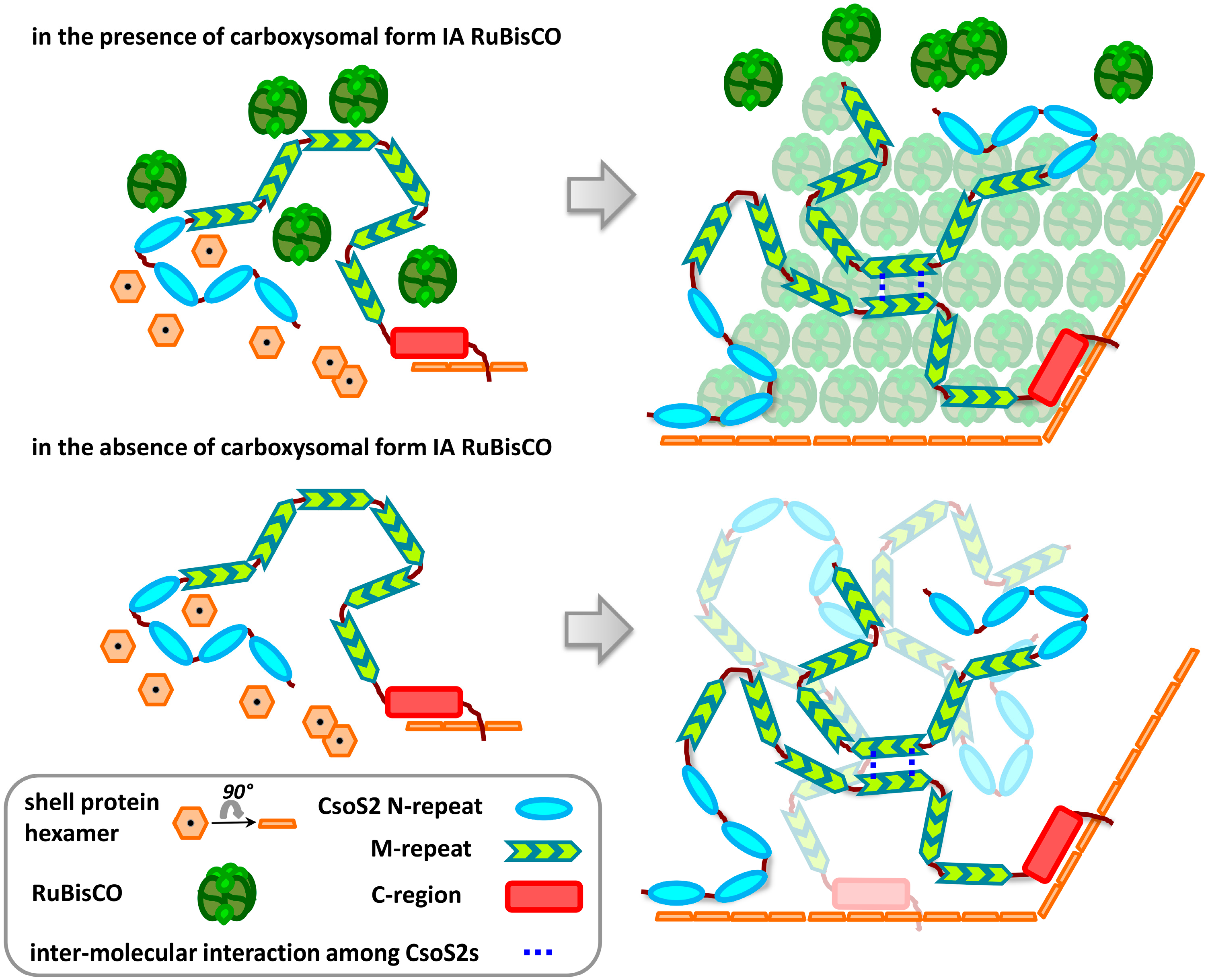
3. Discussion
4. Materials and Methods
4.1. Sequence Analysis and Bioinformatics
4.2. Prediction of Secondary Structure and Protein Folding
4.3. Construction of Hnea csoS2::KmR Mutant
4.4. Construction of HnSPAS2 Mutant
4.5. Purification of HnSPAS2 carboxysomes
4.6. Yeast Two Hybrid Assay
4.7. Characterization of Biophysical Properties
4.8. Peptide Array
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Goericke, R.; Welschmeyer, N.A. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep Sea Res. 1993, 40, 2283–2294. [Google Scholar] [CrossRef]
- Liu, H.; Nolla, H.A.; Campbell, L. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat. Microb. Ecol. 1997, 12, 39–47. [Google Scholar] [CrossRef]
- Liu, H.; Campbell, L.; Landry, M.R.; Nolla, H.A.; Brown, S.L.; Constantinou, J. Prochlorococcus and Synechococcus growth rates and contributions to production in the Arabian Sea during the 1995 Southwest and Northeast Monsoons. Deep Sea Res. 1998, 45, 2327–2352. [Google Scholar] [CrossRef]
- Axen, S.D.; Erbilgin, O.; Kerfeld, C.A. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput. Biol. 2014, 10. [Google Scholar] [CrossRef]
- Marin, B.; Nowack, E.C.; Glockner, G.; Melkonian, M. The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol. Biol. 2007, 7. [Google Scholar] [CrossRef] [PubMed]
- Heinhorst, S.; Cannon, G.C.; Shively, J.M. Carboxysomes and carboxysome-like inclusions. Microbiol. Monogr. 2006, 2, 141–165. [Google Scholar]
- Tanaka, S.; Kerfeld, C.A.; Sawaya, M.R.; Cai, F.; Heinhorst, S.; Cannon, G.C.; Yeates, T.O. Atomic-level models of the bacterial carboxysome shell. Science 2008, 319, 1083–1086. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.W.; Cai, F.; Kerfeld, C.A.; Cannon, G.C.; Heinhorst, S. Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J. Bacteriol. 2012, 194, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, N.M.; Sundberg, C.D.; Gidaniyan, S.D.; Cascio, D.; Yeates, T.O. Structure and identification of a pterin dehydratase-like protein as a RuBisCO assembly factor in the alpha-carboxysome. J. Biol. Chem. 2014, 289, 7973–7981. [Google Scholar] [CrossRef] [PubMed]
- Bonacci, W.; Teng, P.K.; Afonso, B.; Niederholtmeyer, H.; Grob, P.; Silver, P.A.; Savage, D.F. Modularity of a carbon-fixing protein organelle. Proc. Natl. Acad. Sci. USA 2012, 109, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Dou, Z.; Heinhorst, S.; Williams, E.B.; Murin, C.D.; Shively, J.M.; Cannon, G.C. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Bio. Chem. 2008, 283, 10377–10384. [Google Scholar] [CrossRef]
- Menon, B.B.; Dou, Z.; Heinhorst, S.; Shively, J.M.; Cannon, G.C. Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RuBisCO species. PLoS One 2008, 3. [Google Scholar] [CrossRef]
- Menon, B.B.; Heinhorst, S.; Shively, J.M.; Cannon, G.C. The carboxysome shell is permeable to protons. J. Bacteriol. 2010, 192, 5881–5886. [Google Scholar] [CrossRef]
- Cai, F.; Menon, B.B.; Cannon, G.C.; Curry, K.J.; Shively, J.M.; Heinhorst, S. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 2009, 4. [Google Scholar] [CrossRef]
- Klein, M.G.; Zwart, P.; Bagby, S.C.; Cai, F.; Chisholm, S.W.; Heinhorst, S.; Cannon, G.C.; Kerfeld, C.A. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 2009, 392, 319–333. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Cannon, G.C.; Heinhorst, S.; Tanaka, S.; Williams, E.B.; Yeates, T.O.; Kerfeld, C.A. The structure of β-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J. Biol. Chem. 2006, 281, 7546–7555. [Google Scholar] [CrossRef]
- Tsai, Y.; Sawaya, M.R.; Cannon, G.C.; Cai, F.; Williams, E.B.; Heinhorst, S.; Kerfeld, C.A.; Yeates, T.O. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007, 5. [Google Scholar] [CrossRef]
- Tsai, Y.; Sawaya, M.R.; Yeates, T.O. Analysis of lattice-translocation disorder in the layered hexagonal structure of carboxysome shell protein CsoS1C. Acta Crystallogr. 2009, D65, 980–988. [Google Scholar]
- Cai, F.; Sutter, M.; Bernstein, S.L.; Kinney, J.N.; Kerfeld, C.A. Engineering bacterial microcompartment shells: Chimeric shell proteins and chimeric carboxysome shells. ACS Synth. Biol. 2014. [Google Scholar] [CrossRef]
- Dou, Z. Functional characterization and assembly studies of carboxysomes in Halothiobacillus neapolitanus. Ph.D Thesis, The University of Southern Mississippi, Hattiesburg, MS, USA, 2009. [Google Scholar]
- Menon, B.B. Molecular interactions involved in the biogenesis of bacterial microcompartments. Ph.D Thesis, The University of Southern Mississippi, Hattiesburg, MS, USA, 2009. [Google Scholar]
- Baker, S.H.; Lorbach, S.C.; Rodriguez-Buey, M.; Williams, D.S.; Aldrich, H.C.; Shively, J.M. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch. Microbiol. 1999, 172, 233–239. [Google Scholar] [CrossRef]
- Iancu, C.V.; Morris, D.M.; Dou, Z.; Heinhorst, S.; Cannon, G.C.; Jensen, G.J. Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J. Mol. Biol. 2010, 396, 105–117. [Google Scholar] [CrossRef]
- Cameron, J.C.; Wilson, S.C.; Bernstein, S.L.; Kerfeld, C.A. Biogenesis of a bacterial organelle: The carboxysome assembly pathway. Cell 2013, 155, 1131–1140. [Google Scholar] [CrossRef]
- Chien, C.T.; Bartel, P.L.; Sternglanz, R.; Fields, S. The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 1991, 88, 9578–9582. [Google Scholar] [CrossRef]
- So, A.K.-C.; Espie, G.S.; Williams, E.B.; Shively, J.M.; Heinhorst, S.; Cannon, G.C. A novel evolutionary lineage of carbonic anhydrase (ɛ Class) is a component of the carboxysome shell. J. Bacteriol. 2004, 186, 623–630. [Google Scholar] [CrossRef]
- Cannon, G.C.; Baker, S.H.; Soyer, F.; Johnson, D.R.; Bradburne, C.E.; Mehlman, J.L.; Davies, P.S.; Jiang, Q.L.; Heinhorst, S.; Shively, J.M. Organization of carboxysome genes in the Thiobacilli. Curr. Microbiol. 2003, 46, 115–119. [Google Scholar] [CrossRef]
- Ting, C.S.; Dusenbury, K.H.; Pryzant, R.A.; Higgins, K.W.; Pang, C.J.; Black, C.E.; Beauchamp, E.M. The Prochlorococcus carbon dioxide-concentrating mechanism: Evidence of carboxysome-associated heterogeneity. Photosynth. Res. 2015, 123, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hugenholtz, P.; Mavromatis, K.; Pukall, R.; Dalin, E.; Ivanova, N.N.; Kunin, V.; Goodwin, L.; Wu, M.; Tindall, B.J.; et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009, 462, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, L.; Long, B.M.; Price, G.D.; Badger, M.R. Comparing the in vivo function of α-carboxysomes and β-carboxysomes in two model cyanobacteria. Plant Physiol. 2014, 165, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Rahman, F.; Petit, E.; Blanchard, J.L. The distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. J. Phylogenet. Evol. Biol. 2013, 1. [Google Scholar] [CrossRef]
- Di Rienzi, S.C.; Sharon, I.; Wrighton, K.C.; Koren, O.; Hug, L.A.; Thomas, B.C.; Goodrich, J.K.; Bell, J.T.; Spector, T.D.; Banfield, J.F.; et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2013, 2. [Google Scholar] [CrossRef]
- Von Ossowski, I.; Eaton, J.T.; Czjzek, M.; Perkins, S.J.; Frandsen, T.P.; Schulein, M.; Panine, P.; Henrissat, B.; Receveur-Brechot, V. Protein disorder: Conformational distribution of the flexible linker in a chimeric double cellulase. Biophys. J. 2005, 88, 2823–2832. [Google Scholar]
- Hammel, M.; Fierobe, H.P.; Czjzek, M.; Kurkal, V.; Smith, J.C.; Bayer, E.A.; Finet, S.; Receveur-Brechot, V. Structural basis of cellulosome efficiency explored by small angle X-ray scattering. J. Biol. Chem. 2005, 280, 38562–38568. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Zarzycki, J.; Axen, S.D.; Kinney, J.N.; Kerfeld, C.A. Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 2013, 64, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.T.; Occhialini, A.; Andralojc, P.J.; Devonshire, J.; Hines, K.M.; Parry, M.A.; Hanson, M.R. β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant. J. 2014, 79. [Google Scholar] [CrossRef]
- Rae, B.D.; Long, B.M.; Badger, M.R.; Price, G.D. Structural determinants of the outer shell of beta-carboxysomes in Synechococcus elongatus PCC 7942: Roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS One 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Van Eykelenburg, C. Ecophysiological studies on Spirulina platensis. Effect of temperature, light intensity and nitrate concentration on growth and ultrastructure. Antonie Van Leeuwenhoek 1980, 46, 113–127. [Google Scholar]
- Rae, B.D.; Long, B.M.; Badger, M.R.; Price, G.D. Functions, compositions, and evolution of the two types of carboxysomes: Polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 357–379. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Sultemeyer, D.; Price, G.D. Isolation of ccmKLMN genes from the marine cyanobacterium Synechococcus sp. PCC7002 and evidence that CcmM is essential for carboxysome assembly. J. Phycol. 2000, 36, 1109–1118. [Google Scholar] [CrossRef]
- Holthuijzen, Y.A.; VanBreeman, J.F.L.; Konings, W.N.; van Bruggen, E.F.J. Electron microscopic studies of carboxysomes of Thiobacillus neapolitanus. Arch. Microbiol. 1986, 144, 258–262. [Google Scholar] [CrossRef]
- Shively, J.M.; Ball, F.L.; Kline, B.W. Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus. J. Bacteriol. 1973, 116, 1405–1411. [Google Scholar] [PubMed]
- Iancu, C.V.; Ding, H.J.; Morris, D.M.; Dias, D.P.; Gonzales, A.D.; Martino, A.; Jensen, G.J. The Structure of Isolated Synechococcus Strain WH8102 Carboxysomes as Revealed by Electron Cryotomography. J. Mol. Biol. 2007, 372, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.F.; Paredes, A.M.; Khant, H.A.; Soyer, F.; Aldrich, H.C.; Chiu, W.; Shively, J.M. Structure of Halothiobacillus neapolitanus Carboxysomes by Cryo-electron Tomography. J. Mol. Biol. 2006, 364, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, Y.; Danev, R.; Nagayama, K.; Nakamoto, H. Intact carboxysomes in a cyanobacterial cell visualized by hilbert differential contrast transmission electron microscopy. J. Bacteriol. 2006, 188, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Kerfeld, C.A.; Erbilgin, O. Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol. 2015, 23, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Long, B.M.; Tucker, L.; Badger, M.R.; Price, G.D. Functional cyanobacterial beta-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol. 2010, 153, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Cai, F. Transcript analysis of the cso operon and characterization of two sets of conserved bacterial microcompartment genes in the model organism Halothiobacillus neapolitanus. Ph.D. Thesis, The University of Southern Mississippi, Hattiesburg, MS, USA, 2009. [Google Scholar]
- Williams, E.B. Identification and Characterization of Protein Interactions in the Carboxysome of Halothiobacillus neapolitanus. Ph.D. Thesis, The University of Southern Mississippi, Hattiesburg, MS, USA, 2006. [Google Scholar]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. Intrinsically disordered proteins: A 10-year recap. Trends Biochem. Sci. 2012, 37, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, D.; Szabo, B.; Pancsa, R.; Tompa, P. Intrinsically disordered proteins undergo and assist folding transitions in the proteome. Arch. Biochem. Biophys. 2013, 531, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Kinney, J.N.; Salmeen, A.; Cai, F.; Kerfeld, C.A. Elucidating Essential Role of Conserved Carboxysomal Protein CcmN Reveals Common Feature of Bacterial Microcompartment Assembly. J. Biol. Chem. 2012, 287, 17729–17736. [Google Scholar] [CrossRef] [PubMed]
- Long, B.M.; Badger, M.R.; Whitney, S.M.; Price, G.D. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple RuBisCo complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 2007, 282, 29323–29335. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Jakob, U.; Kriwacki, R.; Uversky, V.N. Conditionally and transiently disordered proteins: Awakening cryptic disorder to regulate protein function. Chem. Rev. 2014, 114, 6779–6805. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.M. Intrinsically disordered proteins. Mol. Biosyst. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in scaffold proteins: Getting more from less. Prog. Biophys. Mol. Biol. 2008, 98, 85–106. [Google Scholar] [CrossRef] [PubMed]
- Lassila, J.K.; Bernstein, S.L.; Kinney, J.N.; Axen, S.D.; Kerfeld, C.A. Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J. Mol. Biol. 2014, 426, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers; UCSD Technical Report CS94-351; University of California: San Diego, CA, USA, 1994. [Google Scholar]
- Reimer, U.; Reineke, U.; Schneider-Mergener, J. Peptide arrays: From macro to micro. Curr. Opin. Biotechnol. 2002, 13, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Cretich, M.; Damin, F.; Pirri, G.; Chiari, M. Protein and peptide arrays: Recent trends and new directions. Biomol. Eng. 2006, 23, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Obradovic, Z.; Brown, C.J.; Dunker, A.K. Prediction of boundaries between intrinsically ordered and disordered protein regions. In Proceedings of Pacific Symposium on Biocomputing, Kauai, HI, USA, 3–7 January 2003; pp. 216–227.
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 2005, 61, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Prilusky, J.; Felder, C.E.; Zeev-Ben-Mordehai, T.; Rydberg, E.H.; Man, O.; Beckmann, J.S.; Silman, I.; Sussman, J.L. FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 2005, 21, 3435–3438. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, Y. Toward optimal fragment generations for ab initio protein structure assembly. Proteins 2013, 81, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 2012, 80, 1715–1735. [Google Scholar] [CrossRef] [PubMed]
- Zeghouf, M.; Li, J.; Butland, G.; Borkowska, A.; Canadien, V.; Richards, D.; Beattie, B.; Emili, A.; Greenblatt, J.F. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 2004, 3, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Ellis, H.M.; Lee, E.C.; Jenkins, N.A.; Copeland, N.G.; Court, D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 5978–5983. [Google Scholar] [CrossRef] [PubMed]
- Baker, B. Molecular Investigation of Carbon Dioxide Fixation in Thiobacilli. Ph.D. Thesis, Clemson University, Clemson, SC, USA, 1998. [Google Scholar]
- Baker, S.H.; Jin, S.; Aldrich, H.C.; Howard, G.T.; Shively, J.M. Insertion mutation of the form I cbbL gene encoding ribulose bisphosphate carboxylase/oxygenase (RuBisCO) in Thiobacillus neapolitanus results in expression of form II RuBisCO, loss of carboxysomes, and an increased CO2 requirement for growth. J. Bacteriol. 1998, 180, 4133–4139. [Google Scholar] [PubMed]
- Konarev, P.V.; Petoukhov, M.V.; Volchkov, V.V.; Svergun, D.I. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Cryst. 2006, 39, 277–286. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Svergun, D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Pellois, J.P.; Zhou, X.; Srivannavit, O.; Zhou, T.; Gulari, E.; Gao, X. Individually addressable parallel peptide synthesis on microchips. Nat. Biotechnol. 2002, 20, 922–926. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, F.; Dou, Z.; Bernstein, S.L.; Leverenz, R.; Williams, E.B.; Heinhorst, S.; Shively, J.; Cannon, G.C.; Kerfeld, C.A. Advances in Understanding Carboxysome Assembly in Prochlorococcus and Synechococcus Implicate CsoS2 as a Critical Component. Life 2015, 5, 1141-1171. https://doi.org/10.3390/life5021141
Cai F, Dou Z, Bernstein SL, Leverenz R, Williams EB, Heinhorst S, Shively J, Cannon GC, Kerfeld CA. Advances in Understanding Carboxysome Assembly in Prochlorococcus and Synechococcus Implicate CsoS2 as a Critical Component. Life. 2015; 5(2):1141-1171. https://doi.org/10.3390/life5021141
Chicago/Turabian StyleCai, Fei, Zhicheng Dou, Susan L. Bernstein, Ryan Leverenz, Eric B. Williams, Sabine Heinhorst, Jessup Shively, Gordon C. Cannon, and Cheryl A. Kerfeld. 2015. "Advances in Understanding Carboxysome Assembly in Prochlorococcus and Synechococcus Implicate CsoS2 as a Critical Component" Life 5, no. 2: 1141-1171. https://doi.org/10.3390/life5021141