Highly Iterated Palindromic Sequences (HIPs) and Their Relationship to DNA Methyltransferases
Abstract
:1. Introduction
2. Results
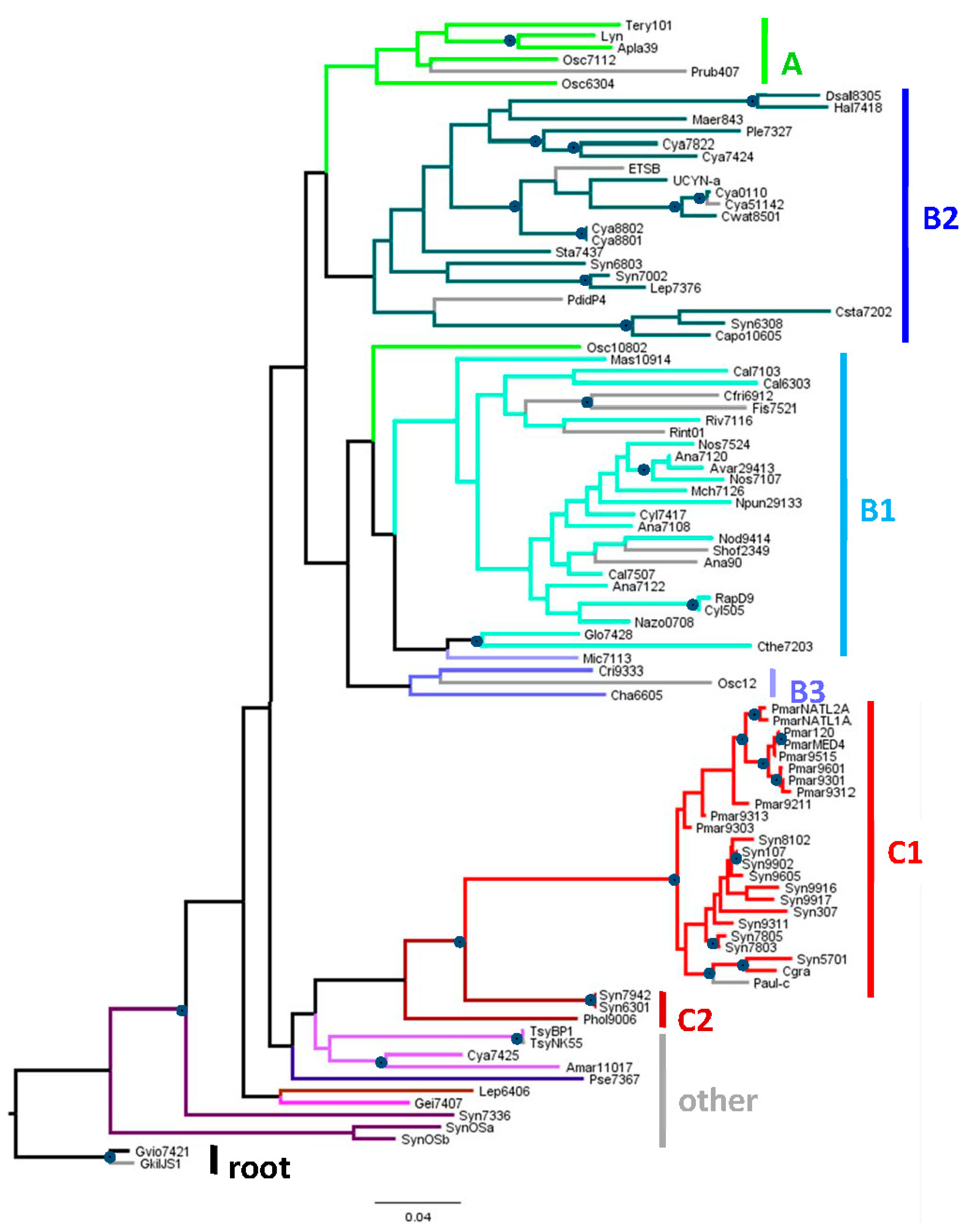
2.1. Genomes Considered in This Study
| Organism | % D253 Fragments Matched | % ID in Matches to Common Fragment |
|---|---|---|
| Calothrix PCC 6303 | 29% | 86% |
| Calothrix PCC 7103 | 79% | 94% |
| Calothrix PCC 7507 | 27% | 83% |
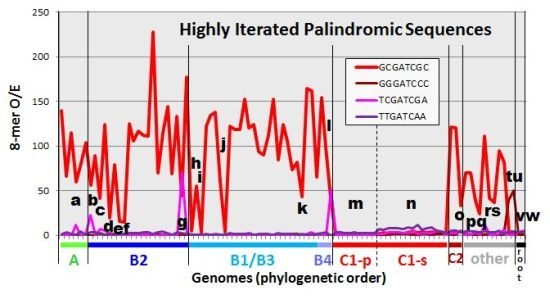
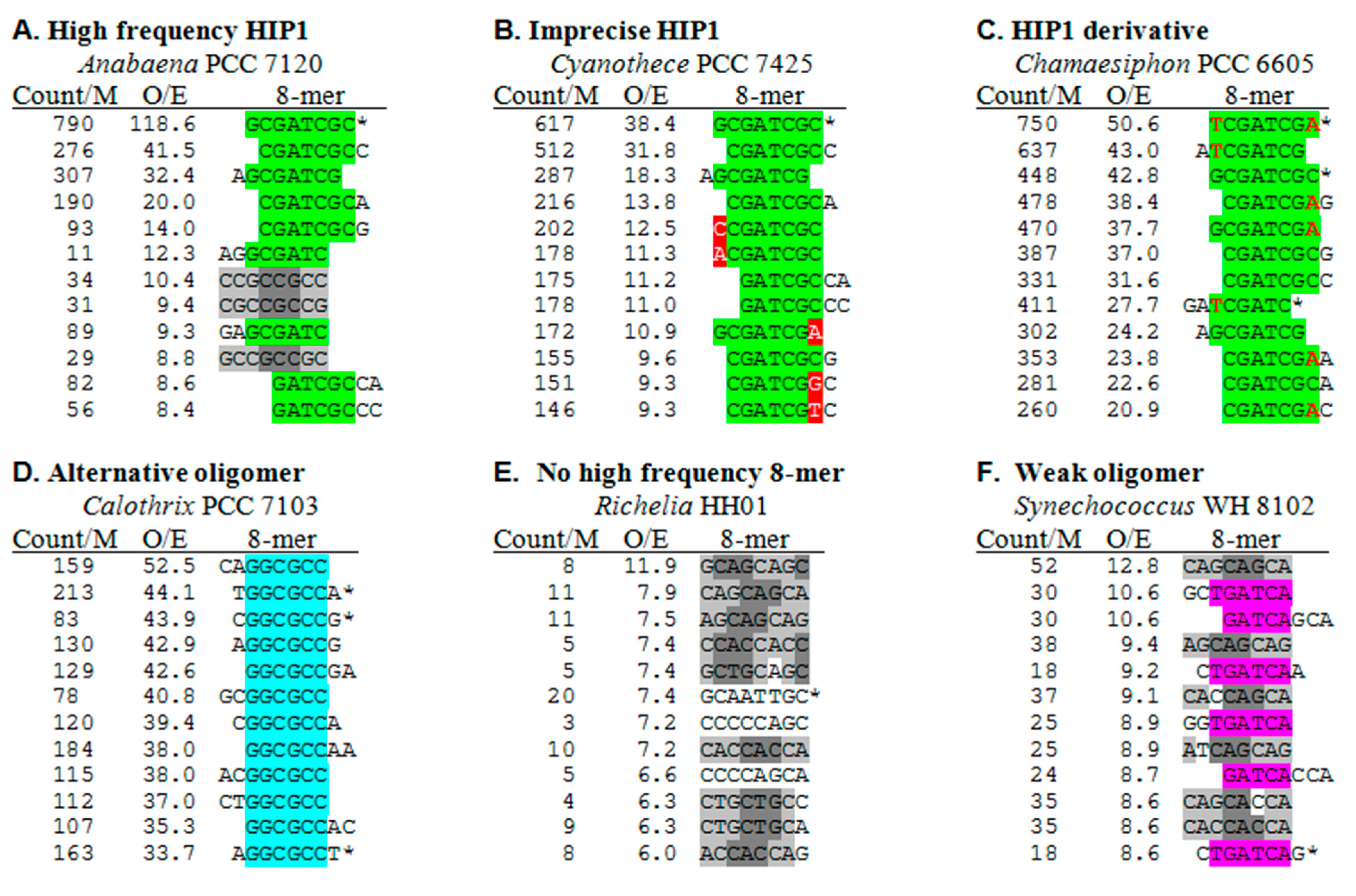
2.2. Frequencies of HIP1 Sequences
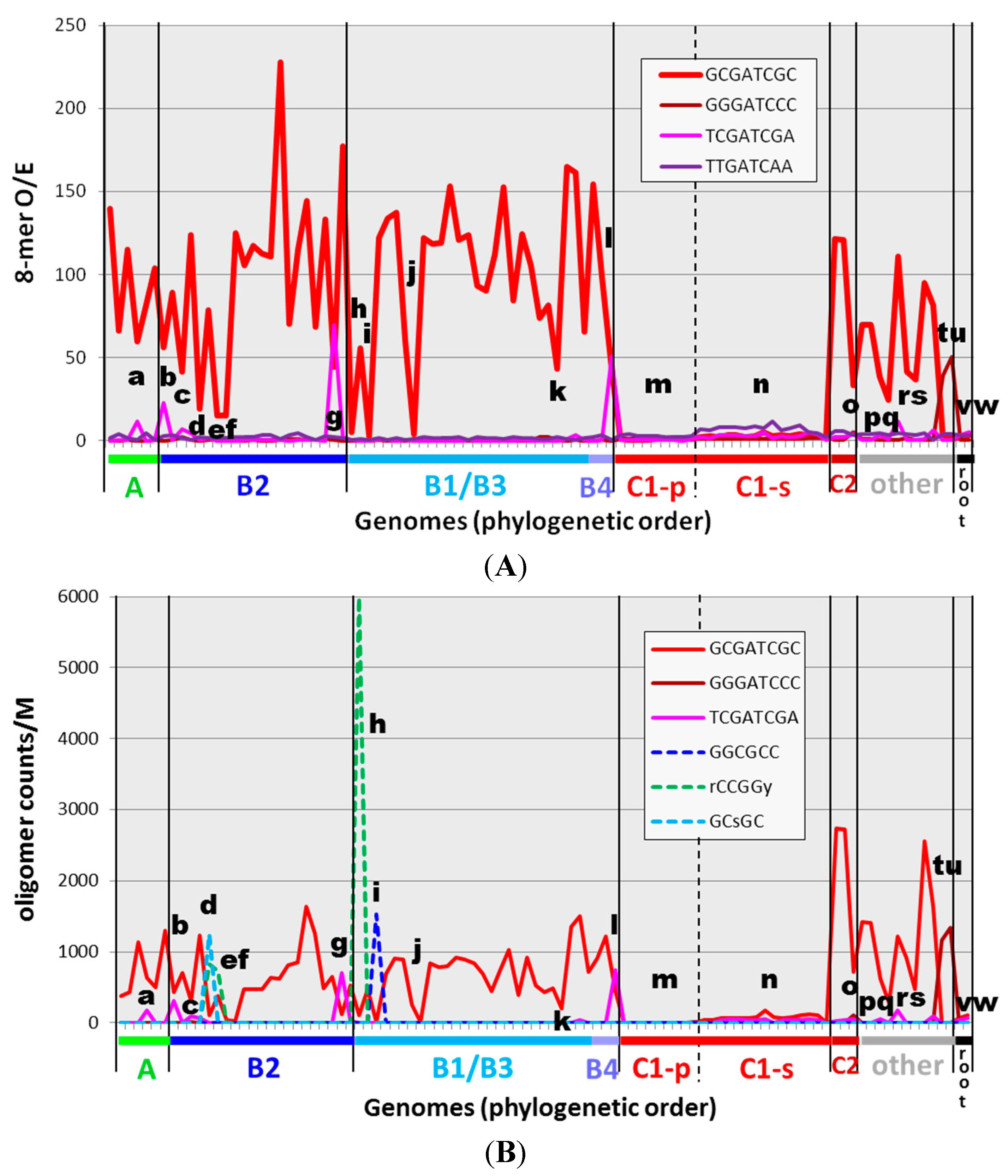
| Organism | HIP1 | Top 8-mer (If Not HIP1) | Comment | ||||
|---|---|---|---|---|---|---|---|
| Count/M | O/E | Sequence | Count/M | O/E | |||
| Most cyanobacteria outside of Group C1 | 300–2700 | 55–228 | - | - | - | High frequency HIP1 | |
| a | A: Oscillatoria PCC 7112 | 630 | 60 | - | - | - | +HIP1 derivative () |
| b | B2: Dactylococcopsis salina PCC 8305 | 425 | 56 | 496 | 66 | +HIP1 derivative () | |
| c | B2: Microcystis aeruginosa NIES 843 | 312 | 42 | - | - | - | Imprecise HIP1 |
| d | B2: Cyanothece PCC 7822 | 111 | 19 | - | - | - | Alternative (GCsGC) |
| e | B2: E.turgida EtSB endosymbiont | 37 | 15 | - | - | - | (Symbiont) Weak imprecise HIP1 |
| f | B2: UCYN-A | 26 | 15 | - | - | - | (Symbiont) Weak HIP1 |
| g | B2: Geminocystis herdmanii PCC 6308 | 121 | 44 | 705 | 70 | HIP1 derivative | |
| h | A: Oscillatoria PCC 10802 | 103 | 5 | 647 | 32 | Alternative (DmtD) | |
| i | B1: Calothrix PCC 7103 | 14 | 3 | 159 | 53 | Alternative () | |
| j | B1: Richellia intracellularis HH01 | 9 | 4 | 30 | 12 | (Symbiont) No high frequency 8-mer | |
| k | B1: Nostoc azollae 0708 | 204 | 43 | - | - | - | (Symbiont) Imprecise HIP1 |
| l | B4: Chamaesiphon minutus PCC 6605 | 448 | 43 | 750 | 51 | HIP1 derivative | |
| m | C1-p: low-GC Prochlorococcus (9) | 1–6 | 0.5–1 | ||||
| n | C1-s: high-GC Prochlorococcus/Synechococcus (15) | 8–171 | 2–5 | various | 52–1278 | 8–21 | Weak oligomer () |
| o | C2: Prochlorothrix hollandica PCC 9006 | 711 | 33 | 753 | 36 | Weak imprecise HIP1 | |
| p | Cyanothece PCC 7425 | 617 | 38 | - | - | - | Weak imprecise HIP1 |
| q | Acaryochloris marina MBIC 11017 | 285 | 24 | - | - | - | Weak HIP1 |
| r | Leptolyngbya PCC 6406 | 925 | 42 | - | - | - | Weak HIP1 |
| s | Leptolyngbya heron island J | 475 | 37 | - | - | - | Weak HIP1 |
| t | Synchococcus OS Type A | 23 | 1 | 1160 | 39 | HIP1 derivative | |
| u | Synechococcus OS Type B | 19 | 1 | 1345 | 50 | HIP1 derivative | |
| v | (root): Gloeobacter violaceus PCC 7421 | 68 | 2 | 43 | 15 | No high frequency 8-mer | |
| w | Gloeobacter kilaueensis JS1 | 102 | 3 | 48 | 14 | No high frequency 8-mer | |
2.3. Frequencies of Other Oligomers
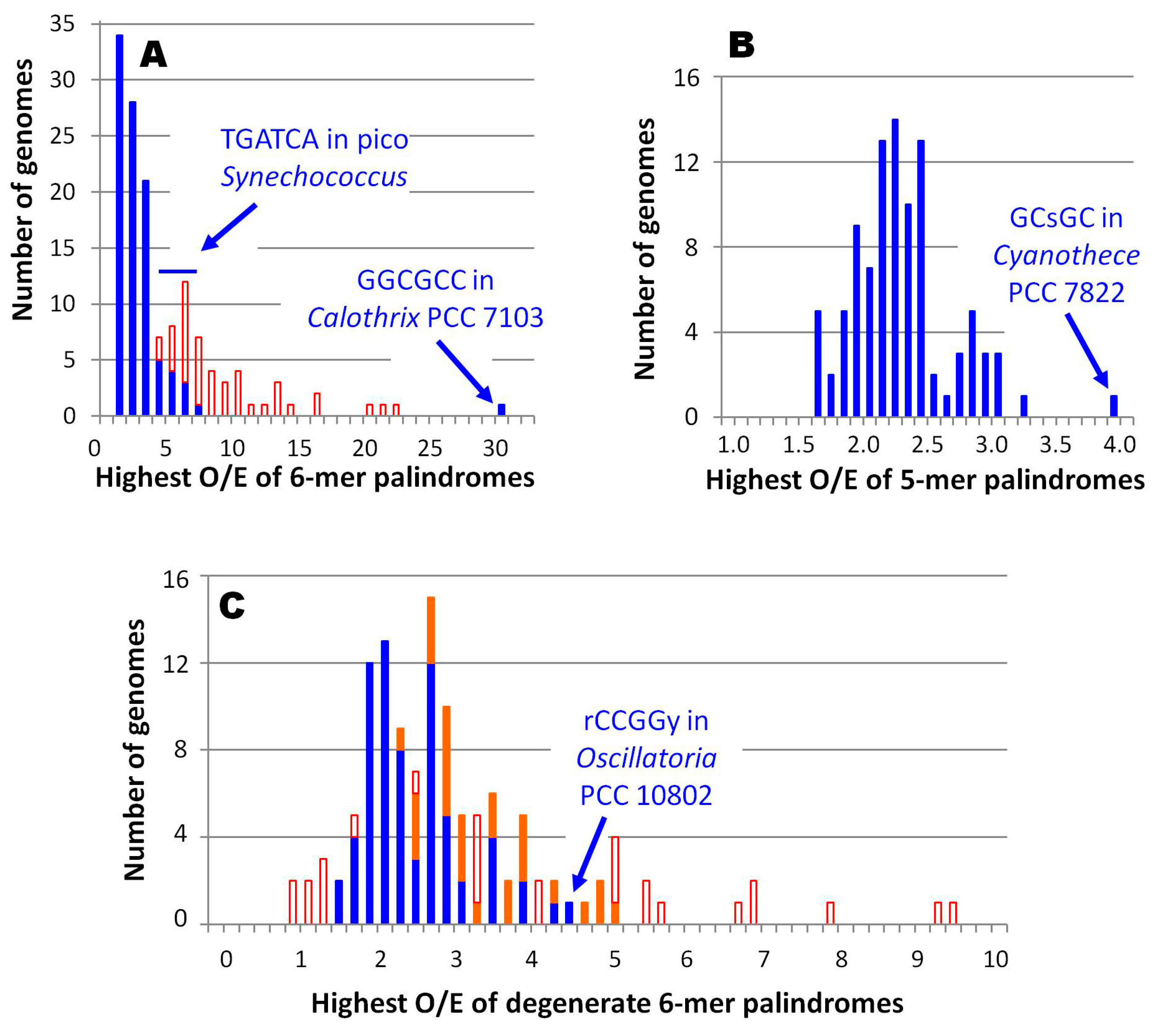
2.4. Extensions to HIP1 and Other High Frequency Oligomers
| Organism | Extended HIP | Extended HIP/HIP |
|---|---|---|
| B2: Cyanobacterium aponimium PCC 10605 | ta | 43% |
| B2: Cyanobacterium stanieri PCC 7202 | gc | 34% |
| B2: Synechocystis PCC 6803 | GC | 88% |
| B2: Leptolyngbya PCC 7376 | GC | 69% |
| B2: Synechococcus PCC 7002 | GC | 71% |
| B2: Dactylococcopsis salina PCC 8305 | gcgc | (30%, 12%) |
| B2: Halothece PCC 7418 | gCGc | (63%, 31%) |
| A: Oscillatoria acuminate PCC 6304 | gc | 27% |
| D: Geitlerinema PCC 7407 | gc | 36% |
| D: Leptolyngbya heron island J | ta | 45% |
| E: Acaryochloris marina MBIC 11017 | TA | 56% |
| C2: Prochlorothrix hollandica PCC 9006 | gc | 30% |
| B1: Oscillatoria PCC 10802 | ca/tg | 33% |
2.5. DNA Methyltransferases Associated with HIP1 and Other Oligomers
2.5.1. GATC Methyltransferases and Their Target Sites
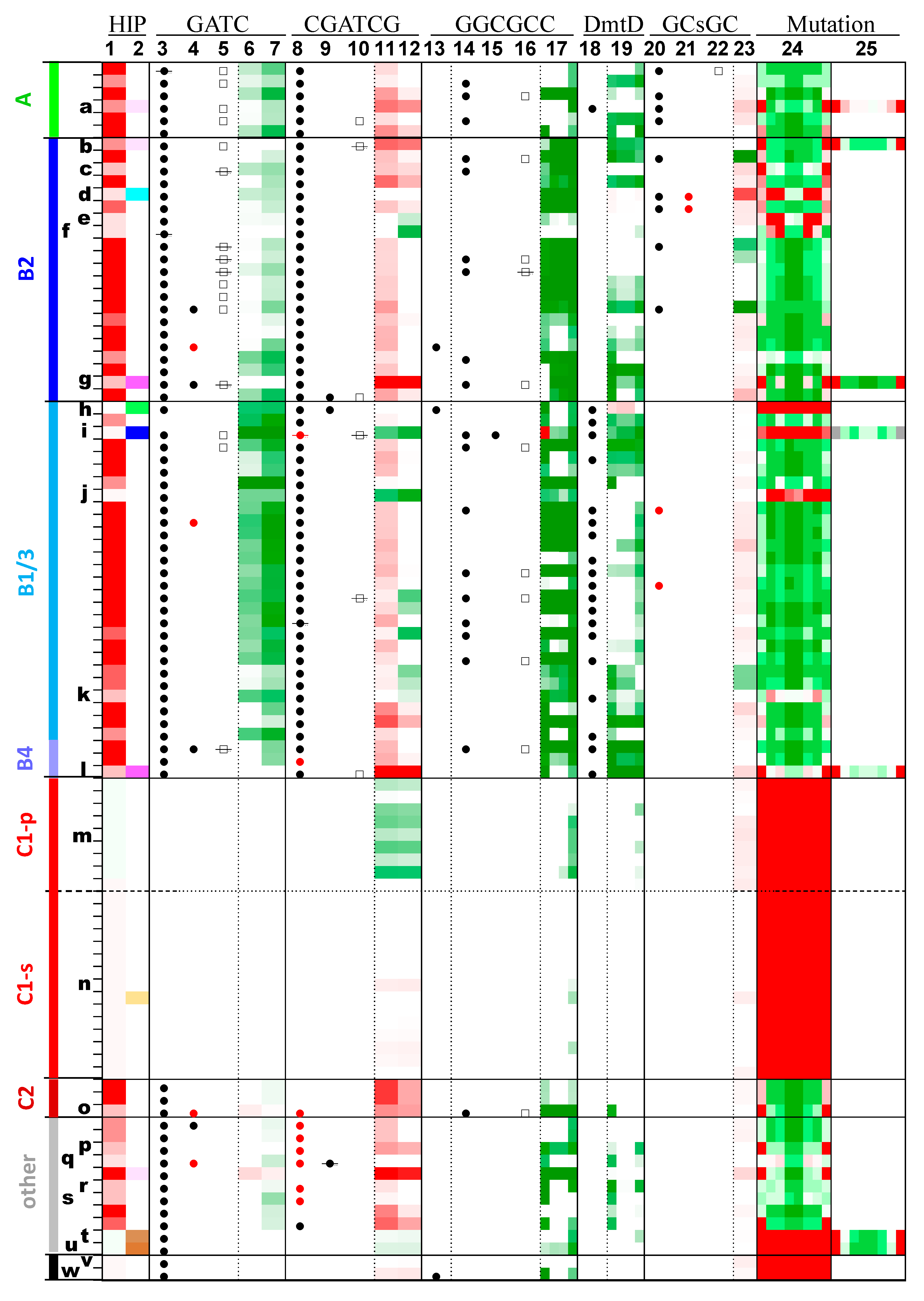
- GATC: (3) GATC-specific MTase, (4) Second GATC-specific MTase, (5) GATC-specific REase, (6) O/E ratio of GATC, (7) O/E ratio of GATC (HIP1 subtracted).
- CGATCG: (8) CGATCG-specific MTase, (9) Second CGATCG-specific MTase, (10) CGATCG-specific REase, (11) O/E ratio of CGATCG, (12) O/E ratio of CGATCG (HIP1 subtracted).
- GGCGCC: (13) GGCGCC-specific MTase, (14) GrCGyC-specific MTase, (15) Second GrCGyC-specific MTase, (16) GrCGyC-specific REase, (17) O/E ratio of sequences specified by GrCGyC: GGCGCC, GGCGTC, GACGCC, GACGTC.
- DmtD: (18) rCCGGy-specific MTase, (19) O/E ratio of sequences specified by rCCGGy: GCCGGC, GCCGGT, ACCGGC, ACCGGT.
- GCsGC: (20) GCsGC-specific MTase, (21) Second GCsGC-specific MTase, (22) GCsGC-specific REase, (23) O/E ratio of GCsGC.
- Mutation: (24) Fraction of sequences deviating from HIP1 at positions 1 through 8, (25) Fraction of sequences deviating from a second highly iterated palindrome (positions 1 through 8), as indicated in Table 2 and Figure 2. For example, the second position is calculated as (CGnGATCGC−CGCGATCGC)/CGnGATCGC, where CS is the counts of the sequence or sequence pattern S, and n represents any nucleotide.
2.5.2. CGATCG Methyltransferases and Their Target Sites
2.5.3. Other Methyltransferases and Their Target Sites
2.6. Substitution Patterns
3. Discussion
3.1. The Nature of HIP Sequences
3.2. The Nature of the Proteins Identified as Methyltransferases
3.3. Functional Roles of Methyltransferases Associated with HIP Sequences
3.4. How are HIP Sequences Gained?
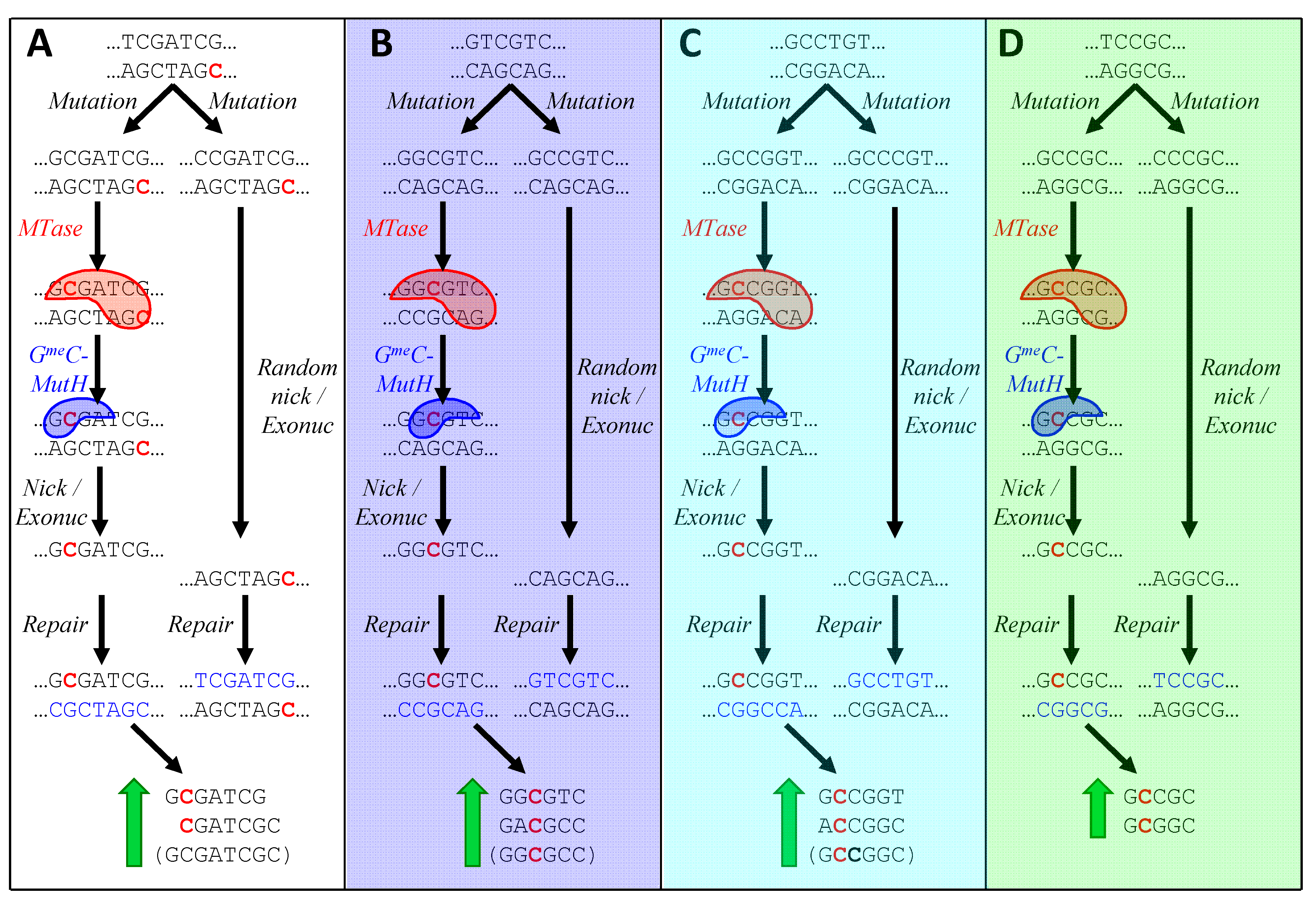
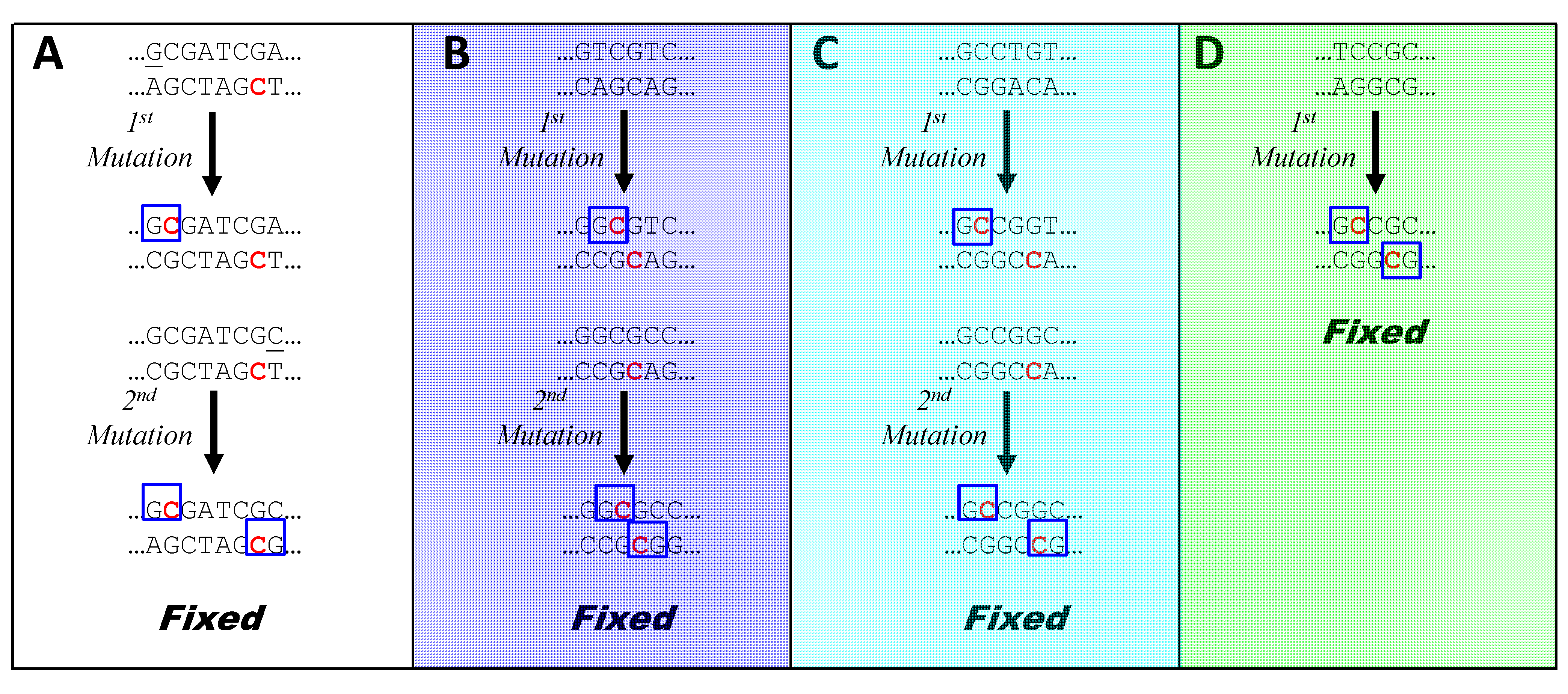
3.5. How is HIP1 Lost? How Are New HIP Sequences Selectively Gained?
3.6. Why Do Symbiotic Cyanobacteria Lose HIP1?
4. Experimental Section
4.1. Phylogenetic Trees
4.2. Protein and Nucleotide Sequences
4.3. Calculation of Occurrences of Oligomer Sequences
- Most deviant red: Cobs/Cexp such that Log10(Pobs/Pexp) = 8000
- Least deviant red: Cobs/Cexp such that Log10(Pobs/Pexp) = 100
- Least deviant green: Cobs/Cexp = Cobs/Cexp for least deviant red
- Most deviant green: Cobs/Cexp = Cobs/Cexp for most deviant red
- Intermediate shades distributed linearly between extreme shades
Acknowledgments
Supplementary Materials
Conflicts of Interest
References
- El Karoui, M.; Biaudet, V.; Schbath, S.; Gruss, A. Characteristics of Chi distribution on different bacterial genomes. Res. Microbiol. 1999, 150, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, O.; Smith, H.O.; Gwinn, M.L.; Salzberg, S.L. DNA uptake signal sequences in naturally transformable bacteria. Res. Microbiol. 1999, 150, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.J.; Robinson, P.J.; Gupta, A.; Bleasby, A.J.; Whitton, B.A.; Morby, A.P. Singular over-representation of an octameric palindrome, HIP1, in DNA from many cyanobacteria. Nucleic Acids Res. 1995, 23, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Delaye, L.; Moya, A. Abundance and distribution of the highly iterated palindrome 1 (HIP1) among prokaryotes. Mob. Genet. Elem. 2011, 1, 159–168. [Google Scholar] [CrossRef]
- Akiyama, H.; Kanai, S.; Hirano, M.; Miyasaka, H. A novel plasmid recombination mechanism of the marine cyanobacterium Synechococcus sp. PCC 7002. DNA Res. 1998, 5, 327–334. [Google Scholar] [CrossRef]
- Robinson, P.J.; Cranenburgh, R.M.; Head, I.M.; Robinson, N.J. HIP1 propagates in cyanobacterial DNA via nucleotide substitutions but promotes excision at similar frequencies in Escherichia coli and Synechococcus PCC 7942. Mol. Microbiol. 1997, 24, 181–189. [Google Scholar] [CrossRef]
- Matveyev, A.V.; Young, K.T.; Meng, A.; Elhai, J. DNA methyltransferases of the cyanobacterium Anabaena PCC 7120. Nucleic Acids Res. 2001, 29, 1491–1506. [Google Scholar] [CrossRef] [PubMed]
- Scharnagl, M.; Richter, S.; Hagemann, M. The cyanobacterium Synechocystis sp. strain PCC 6803 expresses a DNA methyltransferase specific for the recognition sequence of the restriction endonuclease Pvul. J. Bacteriol. 1998, 180, 4116–4122. [Google Scholar]
- Stucken, K.; Koch, R.; Dagan, T. Cyanobacterial defense mechanisms against foreign DNA transfer and their impact on genetic engineering. Biol. Res. 2013, 46, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Elhai, J.; Taton, A.; Massar, J.P.; Myers, J.K.; Travers, M.; Casey, J.; Slupesky, M.; Shrager, J. BioBIKE: A web-based, programmable, integrated biological knowledge base. Nucleic Acids Res. 2009, 37, W28–W32. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; Tandeau de Marsac, N.; Rippka, R.; et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef]
- Saw, J.H.W.; Schatz, M.; Brown, M.V.; Kunkel, D.D.; Foster, J.S.; Shick, H.; Christensen, S.; Hou, S.; Wan, X.; Donachie, S.P.; et al. Cultivation and complete genome sequencing of Gloeobacter kilaueensis sp. nov., from a lava cave in Kılauea Caldera, Hawai’i. PLoS One 2013, 8, e76376. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Ran, L.; Larsson, J.; Vigil-Stenman, T.; Nylander, J.A.; Ininbergs, K.; Zheng, W.W.; Lapidus, A.; Lowry, S.; Haselkorn, R.; Bergman, B. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One 2010, 5, e11486. [Google Scholar] [CrossRef]
- Hilton, J.; Foster, R.; Tripp, H.J.; Carter, B.J.; Zehr, J.P.; Villareal, T.A. Genomic deletions disrupt nitrogen metabolism pathways of a cyanobacterial diatom symbiont. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Thompson, A.W.; Foster, R.A.; Krupke, A.; Carter, B.J.; Musat, N.; Vaulot, D.; Kuypers, M.M.; Zehr, J.P. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 2012, 337, 1546–1550. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Fricke, W.F.; Partensky, F.; Cox, J.; Elshahawi, S.I.; White, J.R.; Phillippy, A.M.; Schatz, M.C.; Piel, J.; Haygood, M.G.; et al. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc. Natl. Acad. Sci. USA 2011, 108, E1423–E1432. [Google Scholar] [CrossRef]
- Nakayama, T.; Kamikawa, R.; Tanifuji, G.; Kashiyama, Y.; Ohkouchi, N.; Archibald, J.M.; Inagaki, Y. Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc. Natl. Acad. Sci. USA 2014, 111, 11407–22412. [Google Scholar] [CrossRef] [PubMed]
- Nowack, E.C.M.; Melkonian, M.; Glöckner, G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 2008, 18, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X. Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 293–318. [Google Scholar] [CrossRef] [PubMed]
- Bakkali, M. Genome dynamics of short oligonucleotides: The example of bacterial DNA uptake enhancing sequences. PLoS One 2007, 2, e741. [Google Scholar] [CrossRef]
- Ahlert, D.; Stegemann, S.; Kahlau, S.; Ruf, S.; Bock, R. Insensitivity of chloroplast gene expression to DNA methylation. Mol. Genet. Genomics 2009, 282, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Barberyron, T.; Kean, K.; Forterre, P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J. Bacteriol. 1984, 160, 586–590. [Google Scholar] [PubMed]
- Padhy, R.N.; Hottat, F.G.; Coene, M.M.; Hoet, P.P. Restriction analysis and quantitative estimation of methylated bases of filamentous and unicellular cyanobacterial DNAs. J. Bacteriol. 1988, 170, 1934–1939. [Google Scholar] [PubMed]
- Murphy, R.C.; Gasparich, G.E.; Bryant, D.A.; Porter, R.D. Nucleotide sequence and further characterization of the Synechococcus sp. strain PCC 7002 recA gene: Complementation of a cyanobacterial recA mutation by the Escherichia coli recA gene. J. Bacteriol. 1990, 172, 967–976. [Google Scholar]
- Domain, F.; Houot, L.; Chauvat, F.; Cassier-Chauvat, C. Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 2004, 53, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Glaeser, J.; Nuss, A.M.; Berghoff, B.A.; Klug, G. Singlet oxygen stress in microorganisms. Adv. Microbiol. Physiol. 2011, 58, 141–173. [Google Scholar] [CrossRef]
- Elhai, J.; Kato, M.; Cousins, S.; Lindblad, P.; Costa, J.L. Very small mobile repeated elements in cyanobacterial genomes. Genome Res. 2008, 18, 1484–1499. [Google Scholar] [CrossRef] [PubMed]
- Marinus, M.G.; Casadesus, J. Roles of DNA adenine methylation in host-pathogen interactions: Mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009, 33, 488–503. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.R.; Pluciennik, A.; Burdett, V.; Modrich, P.L. DNA mismatch repair: Functions and mechanisms. Chem. Rev. 2006, 106, 302–323. [Google Scholar] [CrossRef] [PubMed]
- Guarné, A. The functions of MutL in mismatch repair: The power of multitasking. Prog. Mol. Biol. Transl. 2012, 110, 41–70. [Google Scholar] [CrossRef]
- Bruni, R.; Martin, D.; Jiricny, J. d(GATC) sequences influence Escherichia coli mismatch repair in a distance-dependent manner from positions both upstream and downstream of the mismatch. Nucleic Acids Res. 1988, 16, 4875–4890. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—a database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef]
- Laayoun, A.; Baker, D.J.; Riley, J.; Smith, S.S. The response of M.HpaII to heteroduplexes. Gene 1994, 150, 195–196. [Google Scholar] [CrossRef]
- Renbaum, P.; Razin, A. Interaction of M.SssI and M.HhaI with single-base mismatched oligodeoxynucleotide duplexes. Gene 1995, 157, 177–179. [Google Scholar] [CrossRef]
- Rusmintratip, V.; Riggs, A.D.; Sowers, L.C. Examination of the DNA substrate selectivity of DNA cytosine methyltransferases using mass tagging. Nucleic Acids Res. 2000, 28, 3594–3599. [Google Scholar] [CrossRef] [PubMed]
- Langhans, M.T.; Palladino, M.J. Cleavage of mispaired heteroduplex DNA substrates by numerous restriction enzymes. Curr. Issues Mol. Biol. 2009, 11, 1–12. [Google Scholar] [PubMed]
- Rocha, E.P.; Danchin, A.; Viari, A. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 2001, 11, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Elhai, J. Determination of bias in the relative abundance of oligonucleotides in DNA sequences. J. Comput. Biol. 2001, 8, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhang, X.; Liang, C.; Wu, J.; Bao, Q.; Qin, S. Genome-wide analysis of restriction-modification system in unicellular and filamentous cyanobacteria. Physiol. Genomics 2006, 24, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Budroni, S.; Siena, E.; Hotopp, J.C.D.; Seib, K.L.; Serruto, D.; Nofroni, C.; Comanducci, M.; Riley, D.R.; Daugherty, S.C.; Angiuoli, S.V.; et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc. Natl. Acad. Sci. USA 2011, 108, 4494–4499. [Google Scholar] [CrossRef]
- Krieger-Liszkay, A. Singlet oxygen production in photosynthesis. J. Exp. Bot. 2005, 56, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Triantaphylidès, C.; Krischke, M.; Hoeberichts, F.A.; Ksas, B.; Gresser, G.; Havaux, M.; van Breusegem, F.; Mueller, M.J. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008, 148, 960–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GuhaMajumdar, M.; Sears, B.B. Chloroplast DNA base substitutions: An experimental assessment. Mol. Gen. Genomics 2005, 273, 177–183. [Google Scholar] [CrossRef]
- Wyrzykowski, J.; Volkert, M.R. The Escherichia coli methyl-directed mismatch repair system repairs base pairs containing oxidative lesions. J. Bacteriol. 2003, 185, 1701–1704. [Google Scholar] [CrossRef] [PubMed]
- Golyasnaya, N.V.; Tsvetkova, N.A. Mismatch repair. Mol. Biol. 2006, 40, 183–193. [Google Scholar] [CrossRef]
- Hilton, J.; Satinsky, B.M.; Doherty, M.; Zielinski, B.; Zehr, J.P. Metatranscriptomics of N2-fixing cyanobacteria in the Amazon River plume. ISME J. 2014. [Google Scholar] [CrossRef]
- Peters, G.A.; Meeks, J.C. The Azolla-Anabaena symbiosis: Basic biology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 193–210. [Google Scholar] [CrossRef]
- Griese, M.; Lange, C.; Soppa, J. Ploidy in cyanobacteria. FEMS Microbiol. Lett. 2011, 323, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Rambaud, A. Fig Tree. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 10 March 2015).
- Felsenstein, J. PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 1989, 5, 164–166. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Malone, T.; Blumenthal, R.M.; Cheng, X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995, 253, 618–632. [Google Scholar] [CrossRef] [PubMed]
- Murray, I.A.; Clark, T.A.; Morgan, R.D.; Boitano, M.; Anton, B.P.; Luong, K.; Fomenkov, A.; Turner, S.W.; Korlach, J.; Roberts, R.J.; et al. The methylomes of six bacteria. Nucleic Acids. Res. 2012, 40, 11450–11462. [Google Scholar] [CrossRef]
- Posfai, J.; New England BioLabs, Ipswich, MA, USA. Personal Communication, 2014.
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elhai, J. Highly Iterated Palindromic Sequences (HIPs) and Their Relationship to DNA Methyltransferases. Life 2015, 5, 921-948. https://doi.org/10.3390/life5010921
Elhai J. Highly Iterated Palindromic Sequences (HIPs) and Their Relationship to DNA Methyltransferases. Life. 2015; 5(1):921-948. https://doi.org/10.3390/life5010921
Chicago/Turabian StyleElhai, Jeff. 2015. "Highly Iterated Palindromic Sequences (HIPs) and Their Relationship to DNA Methyltransferases" Life 5, no. 1: 921-948. https://doi.org/10.3390/life5010921