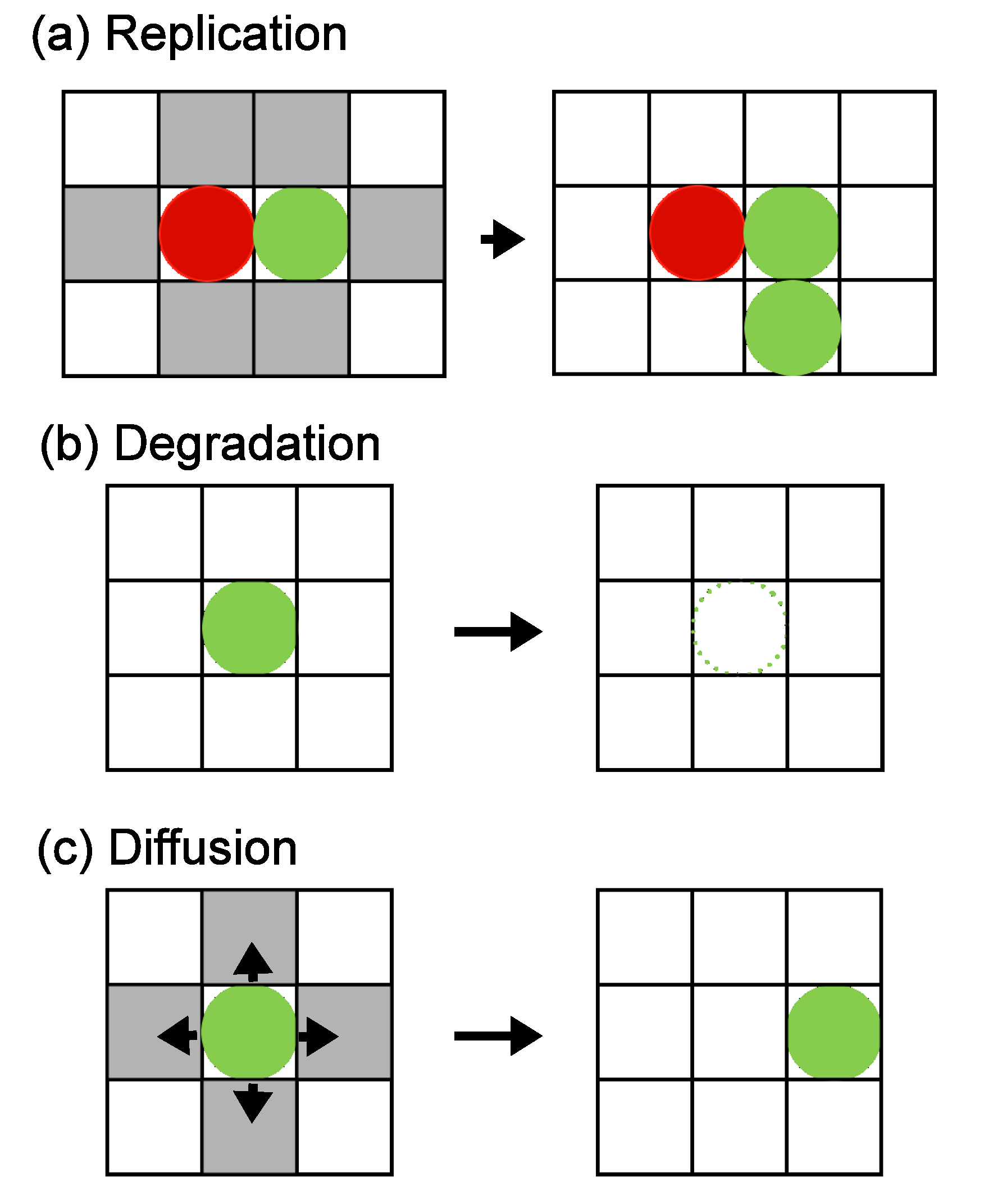
3.1. Two Mutually Catalyzing Molecules
The simplest hypercycle is the case of two molecular species
X and
Y that mutually catalyze the replications as follows:
When
X and
Y are located next to each other, replication can occur with probability
p, as explained in Section 2. If an empty site is available, a new molecule is added, and the molecular species is assigned as
X or
Y , with probabilities
γX and
γY = 1
− γX, respectively. Here we denote
pY =
pγY and
pX =
pγX . Degradations occur as
X → 0 or
Y → 0 with degradation probabilities
aX and
aY, respectively.
Simulations are carried out from the initial condition in which a single Y is located in a group of X with dimensions Lini × Lini. Here, the value of Lini is fixed to 10; however, the exact dimensions are not important if they are sufficiently large.
When the replication and degradation rates for
Y are considerably slower than those for
X, the mutually catalytic replication reactions result in the emergence of a localized spatial structure. This agrees with the structure reported in [
22], in which a single
Y is surrounded by a group of
X (
Figure 2a).
In addition, division of the structure is observed synchronously with the replication of
Y . As
Y replicates at a slow rate, the two
Y molecules start to diffuse apart (
Figure 2a). As each
Y catalyzes the replications of
X, several replicated
X molecules crowd around each of
Y . The size of each structure is determined by the average distance over which
X molecules diffuse before they degrade. Then, when
aY is much smaller than
aX, the two
Y molecules diffuse over a larger distance than the size of the cluster, forming a dumbbell-like cluster around the
Y molecules and resulting in division of the cluster. Through division after the replication of
Y , the number of
X molecules also doubles (
Figure 2b), and recursive growth by successive replications of
Y molecules is possible. Here, we also note that this lattice model allows for a case in which the distance between the two
Y molecules decreases again through diffusion, leading to a decrease in the number of
X molecules as the effective replication rate of
X decreases.
The division process is enabled by the synergetic effect of molecular discreteness and crowding. Shnerb
et al. [
17,
18] investigated a system with a reaction
X +
Y → 2
X +
Y and showed that the discrete number of
Y molecules leads to localized subpopulations and results in a proliferating phase in which the number of
X molecules approaches infinity, even for parameter ranges in which the corresponding reaction-diffusion equation leads to extinction of
X.
Figure 2.
Division of a compartmentalized structure for the model of two mutually catalytic molecules X (green) and Y (red). Parameters are pY = pγY = 5 × 10−6, pX = 1 − pY, aX = 0.01, aY = 0, and Lx = Ly = 1000. (a) Snapshots of the system. Y molecules are highlighted by arrows. The time steps increase from top left to top right, and then from bottom left to bottom right. Snapshots are shown for every 500 steps from 25,000 to 27,500. (b) Time evolution of the number of X and Y molecules corresponding to the data of (a).
Figure 2.
Division of a compartmentalized structure for the model of two mutually catalytic molecules X (green) and Y (red). Parameters are pY = pγY = 5 × 10−6, pX = 1 − pY, aX = 0.01, aY = 0, and Lx = Ly = 1000. (a) Snapshots of the system. Y molecules are highlighted by arrows. The time steps increase from top left to top right, and then from bottom left to bottom right. Snapshots are shown for every 500 steps from 25,000 to 27,500. (b) Time evolution of the number of X and Y molecules corresponding to the data of (a).
In our model, the realistic molecular exclusion volume effect introduces a feedback effect in which crowded
X molecules suppress molecular replications because at most four
X molecules can react with the single
Y in each step and the crowded molecules result in a lack of space near the reacting pair. Thus, a localized structure is formed as a “quasi-stationary” state, in which the number of
X molecules fluctuates around an average value unless
Y replicates or degrades. As
Y replicates, the number of
X molecules shows a step-like increase (
Figure 2b).
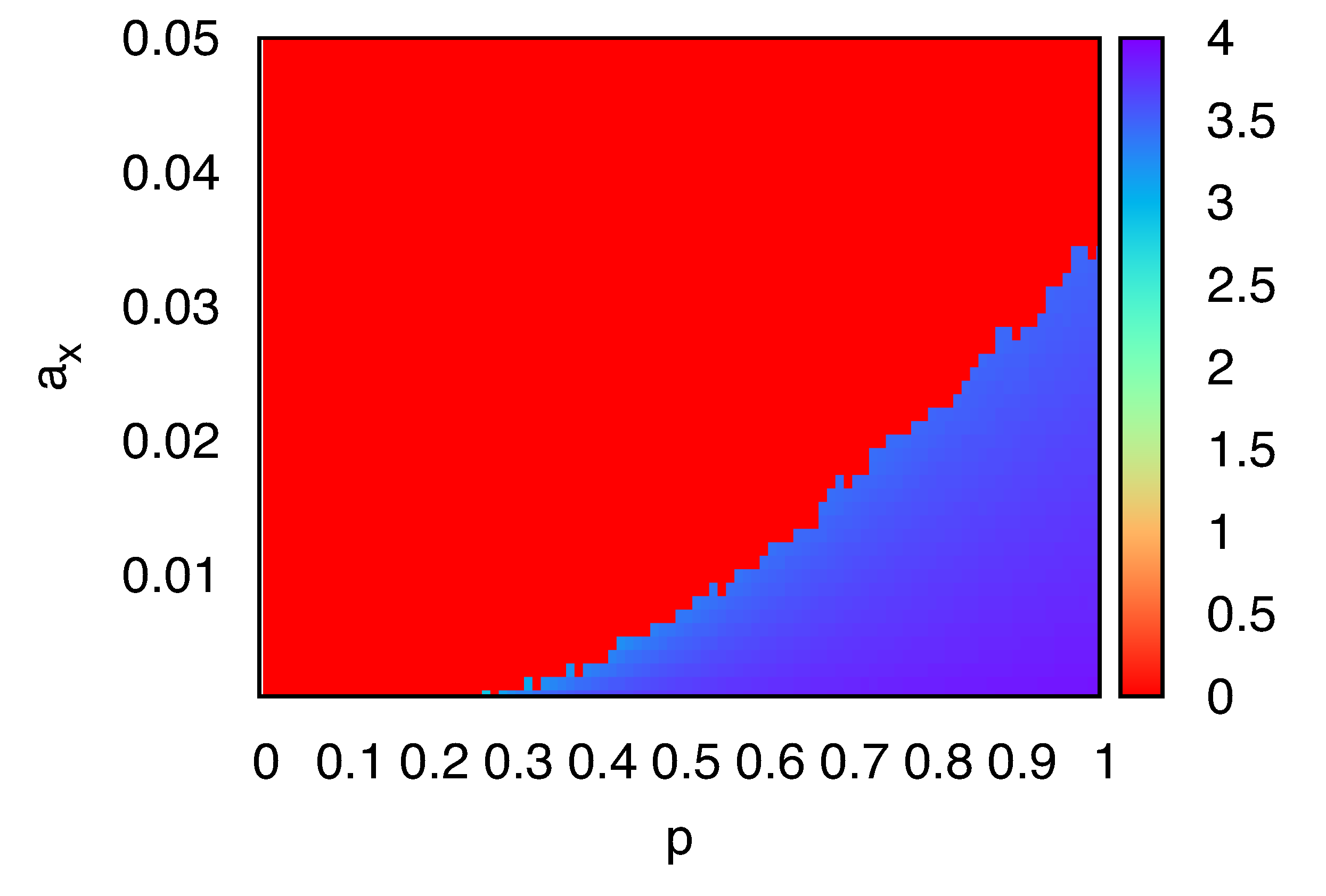
To further clarify the impact of molecular crowding, we investigated the density of
X molecules in the sites neighboring the single
Y. The average number of
X,
nX, in the four nearest-neighboring sites of the single
Y is shown in
Figure 3, as a function of
p and
aX corrected for the change in number of
Y molecules,
i.e., replication/degradation of
Y are absent in computation of the average crowdedness. There are two distinct planes
nX = 0 and
nX > 3.5, against the parameters
p and
aX . In the region
nX = 0, no replication occurs and degradation of
X leads to extinction. In the region
nX > 3.5, localized structure appears. We note that, in simulations with parameters near the boundary between the two regions, the behavior of the system depends on samples: the localized stationary structure or extinction. As long as the system maintains the structure, the value of
nX is approximately the fully-occupied value 4. These results suggest that the discreteness of
Y molecules and the exclusion volume effect set an upper limit for the effective replication rate of
X. If the degradation rate of
X is smaller than the replication rate, replication of
X results in the crowding of
X around the single
Y . If the degradation rate of
X is greater than the replication rate, extinction of
X is observed.
Figure 3.
The average number of X, nX, in the four nearest-neighboring sites around a single Y molecule, plotted as a function of p and aX. Here, γY = aY = 0.
Figure 3.
The average number of X, nX, in the four nearest-neighboring sites around a single Y molecule, plotted as a function of p and aX. Here, γY = aY = 0.
Depending on the parameters
aX,
pX,
aY and
pY, we observe three types of behavior for the
pY-
aY plane: extinction, division and explosion (
Figure 4). The division process is observed for smaller
pY and
aY in the region of
Figure 4 designated by red circles. For larger
pY (explosion region; green squares), both molecules grow and maintain a single cluster with mixed configuration (
Figure 4, right). For larger
aY (extinction; blue diamonds),
Y degradation is followed by the degradation of all
X molecules, resulting in the extinction of all molecules.
Figure 4.
Three types of behavior from the initial condition in the pY-aY space. Here, pY = pγY is changed by γY < 0.5 with p fixed to one. pX = 1 − pY and aX = 0.01. The plane is divided into three regions: extinction (blue diamonds), division (red circles) and explosion (green squares). Typical snapshots are also shown for the division and explosion regions. In the left snapshot, the two existing Y molecules (red) are highlighted by arrows.
Figure 4.
Three types of behavior from the initial condition in the pY-aY space. Here, pY = pγY is changed by γY < 0.5 with p fixed to one. pX = 1 − pY and aX = 0.01. The plane is divided into three regions: extinction (blue diamonds), division (red circles) and explosion (green squares). Typical snapshots are also shown for the division and explosion regions. In the left snapshot, the two existing Y molecules (red) are highlighted by arrows.
3.2. Three Cyclically Catalyzing Molecules
After the two-species system, the next simplest case is a hypercycle consisting of three molecular species (
X,
Y and
Z) that catalyze the replications of other species cyclically as follows:
Here, we denote the replication probabilities of X, Y , and Z by pX, pY, and pZ, respectively. Degradations also occur as X → 0, Y → 0 and Z → 0 with degradation probabilities aX, aY and aZ, respectively.
Simulations are carried out from the initial condition in which a single Y is located in a box with dimensions Lini × Lini with randomly located X and Z molecules.
For a specific set of parameters, the system organizes a localized nested structure in which a single
Y is surrounded by several
X molecules, which are surrounded by a number of
Z molecules (
Figure 5, top left panel). When the replication rate of
Y is small, a cluster of
X molecules is distributed around
Y because
Y catalyzes the replication of
X.
X, in turn, catalyzes the replication of
Z such that several
Z molecules are distributed around the cluster of
X molecules. Replication of
Y is catalyzed by
Z, which is generally separated from the
Y molecule. When a
Z molecule diffuses into the cluster of
X molecules surrounding
Y , a new
Y molecule can be replicated. After replication, the two
Y molecules diffuse apart, and the structure gradually divides (
Figure 5). The replication of
Y also doubles the numbers of
X and
Z molecules, and recursive growth by successive replications of
Y is possible (
Figure 6).
Figure 5.
Snapshots of the system. The X, Y and Z molecules are respectively denoted by green, red and blue dots. The Y molecules are highlighted with arrows. The time steps increase from top left to top right, and then from bottom left to bottom right. Snapshots for every 1000 steps are shown.
Figure 5.
Snapshots of the system. The X, Y and Z molecules are respectively denoted by green, red and blue dots. The Y molecules are highlighted with arrows. The time steps increase from top left to top right, and then from bottom left to bottom right. Snapshots for every 1000 steps are shown.
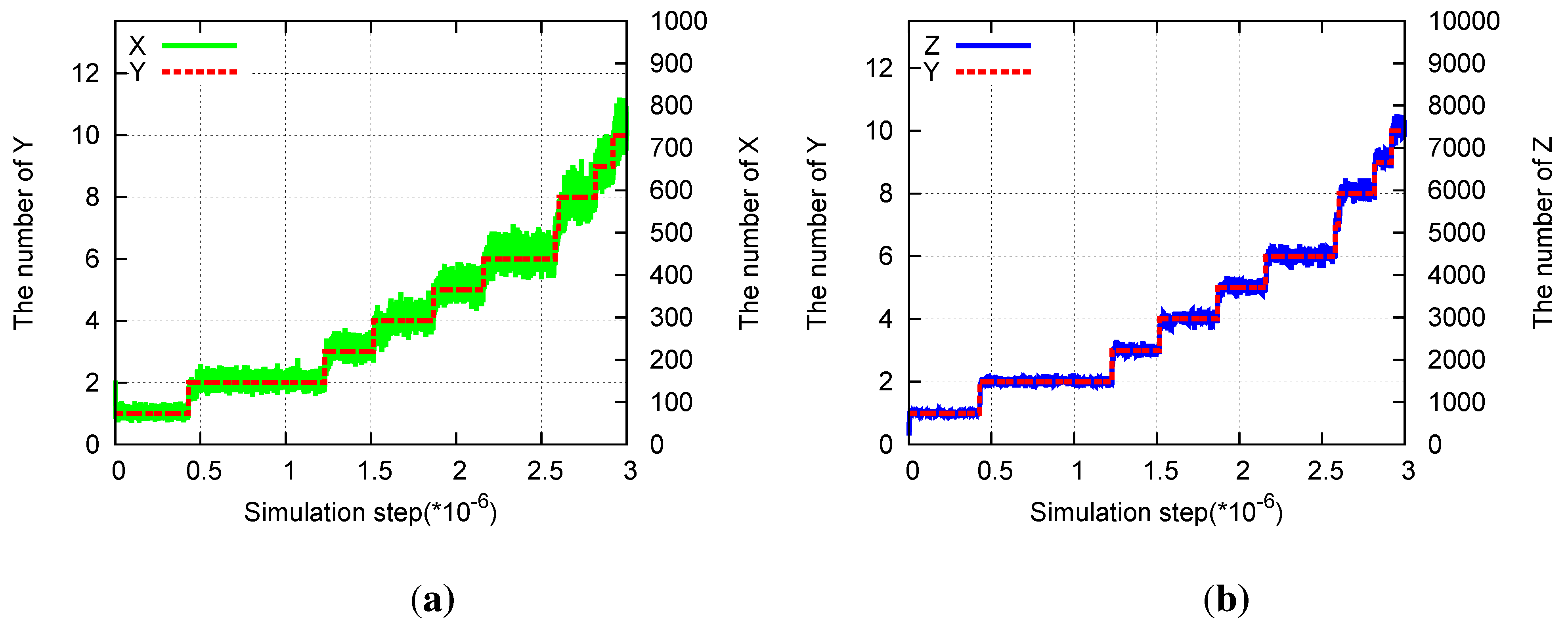
Figure 6.
Division in the three-species hypercycle. Parameters are pX = 1, pY = 0.0001, pZ = 0.01, aX = 0.01, aY = 0, aZ = 0.001. (a) Time evolution of the number of X(green) and of Y (red) molecules. (b) Time evolution of the number of Z(blue) and of Y (red) molecules.
Figure 6.
Division in the three-species hypercycle. Parameters are pX = 1, pY = 0.0001, pZ = 0.01, aX = 0.01, aY = 0, aZ = 0.001. (a) Time evolution of the number of X(green) and of Y (red) molecules. (b) Time evolution of the number of Z(blue) and of Y (red) molecules.
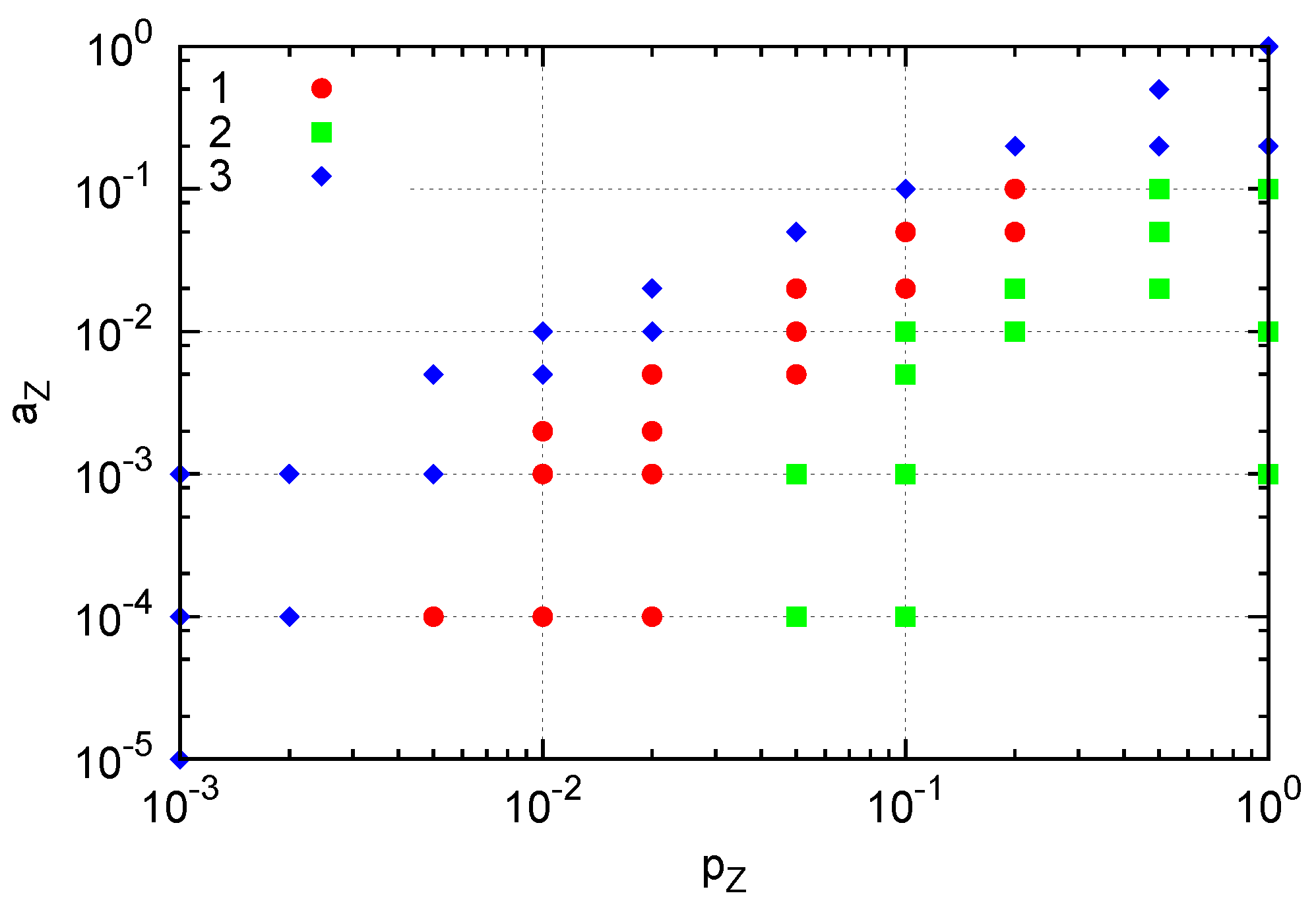
In our model, new molecules are replicated when an empty site is available near the reacting pair. Hence the effective replication rate depends on the spatial distribution of molecules:
i.e., replications are suppressed in crowded situations. With three species, a pair of reacting species is more difficult to interact than in the two-species case. Still, a localized cluster sustaining replication is formed in a certain range of parameters. The parameter region of
pZ and
aZ in which the localized structure appears when a single
Y molecule is fixed is indicated by red circles in
Figure 7. For smaller
pZ or greater
aZ values (blue diamonds), extinction of
Z occurs and prevents the growth of the structure. On the other hand, greater
pZ values (green squares) yield crowded
Z molecules, effectively preventing replication of
X and resulting in the extinction of
X and subsequently extinction of
Z. This result indicates that formation of a dividing cluster crucially depends on the relative replication and degradation rates between
Y and the other species as in the two-species case, but also between
X and
Z molecules in this three-species case. In the parameter space of
Figure 7, for example,
pZ should be smaller than
pX for the emergence of the cluster.
Figure 7.
Three types of generated structures. A single Y molecule is fixed by suppressing changes in Y molecule (i.e., pY = aY = 0). (1) A cluster of Z molecules surrounding a cluster of X molecules around a single Y (red). (2) Extinction of both X and Z: crowding of Z results in extinction of X, and subsequent extinction of Z. (3) Extinction of Z occurs. Here, pX = 1, pY = 0, aX = 0.01 and aY = 0.
Figure 7.
Three types of generated structures. A single Y molecule is fixed by suppressing changes in Y molecule (i.e., pY = aY = 0). (1) A cluster of Z molecules surrounding a cluster of X molecules around a single Y (red). (2) Extinction of both X and Z: crowding of Z results in extinction of X, and subsequent extinction of Z. (3) Extinction of Z occurs. Here, pX = 1, pY = 0, aX = 0.01 and aY = 0.