Synergism and Mutualism in Non-Enzymatic RNA Polymerization
Abstract
:1. Introduction
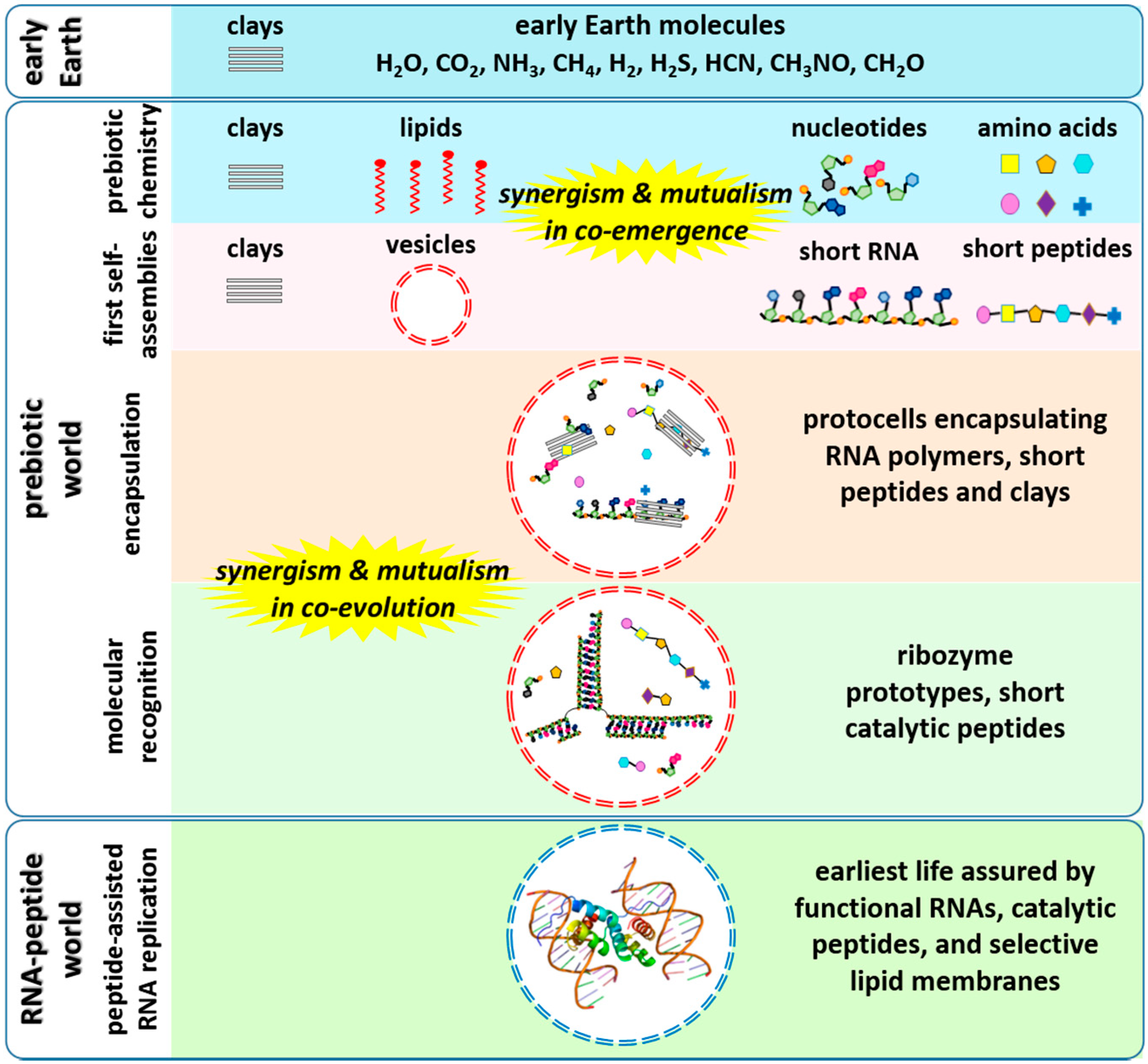
2. Non-Enzymatic RNA Polymerization
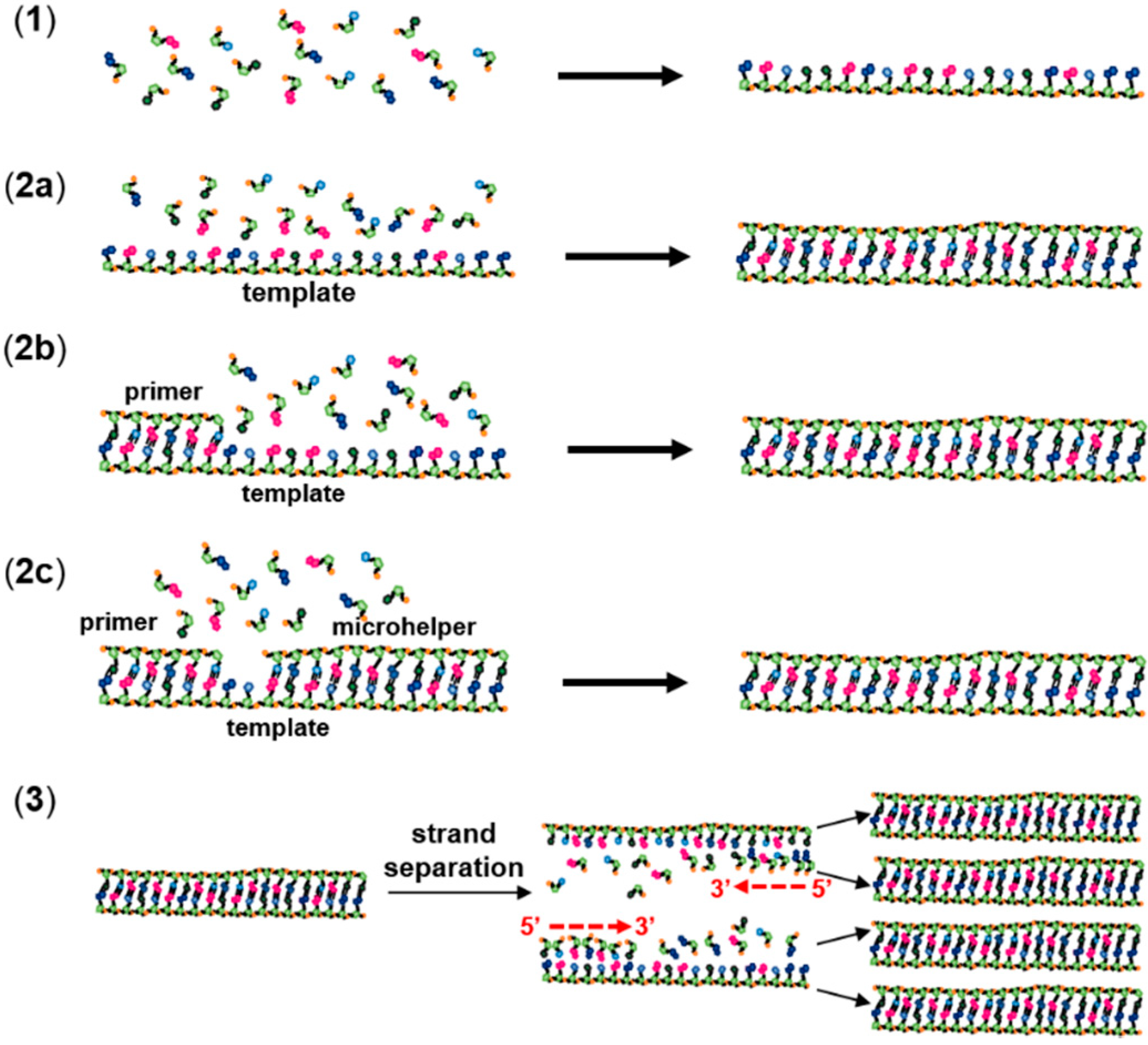
3. The RNA World Hypothesis
4. Thermodynamics of RNA Polymerization
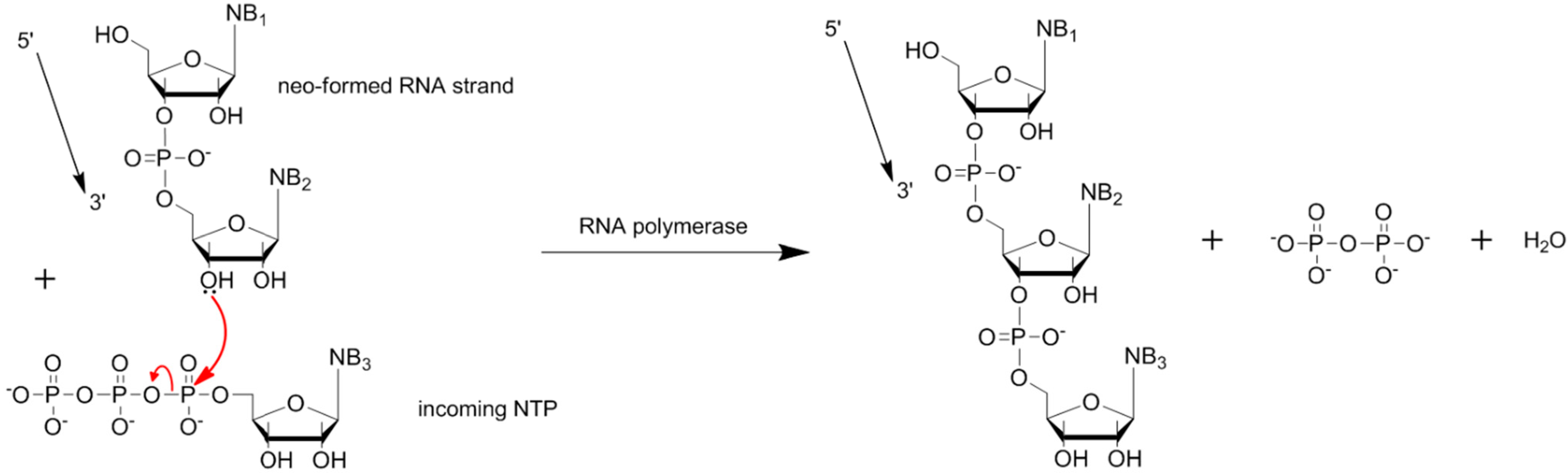
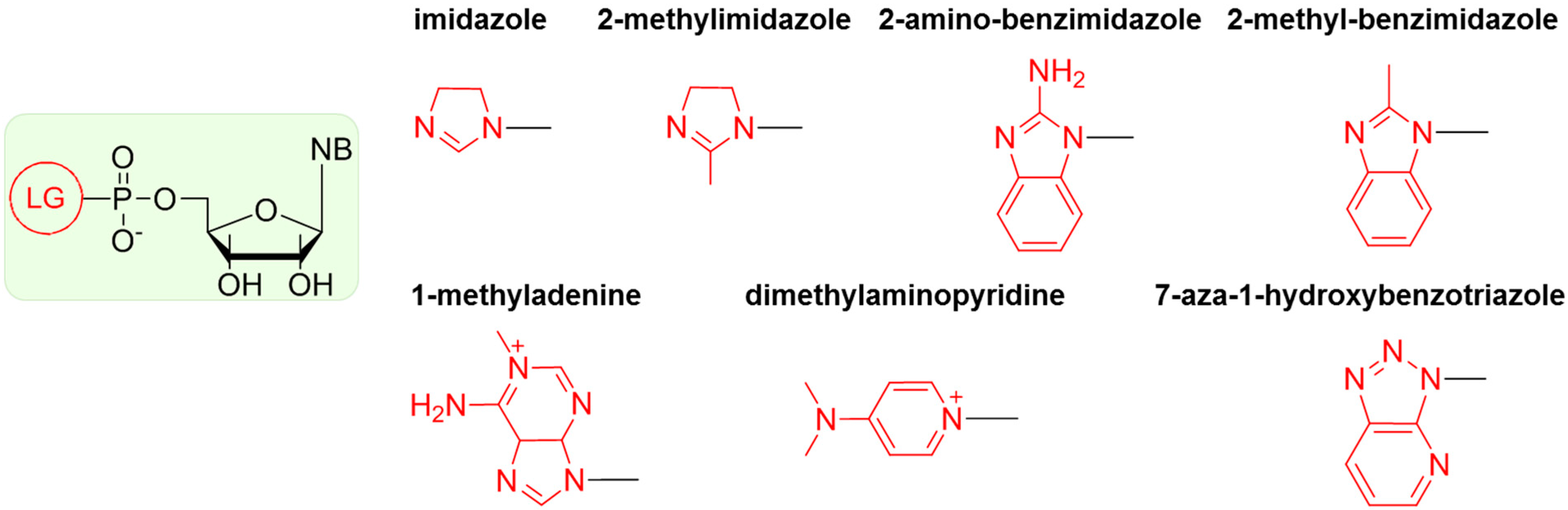
5. Chemical Activation of Mononucleotides
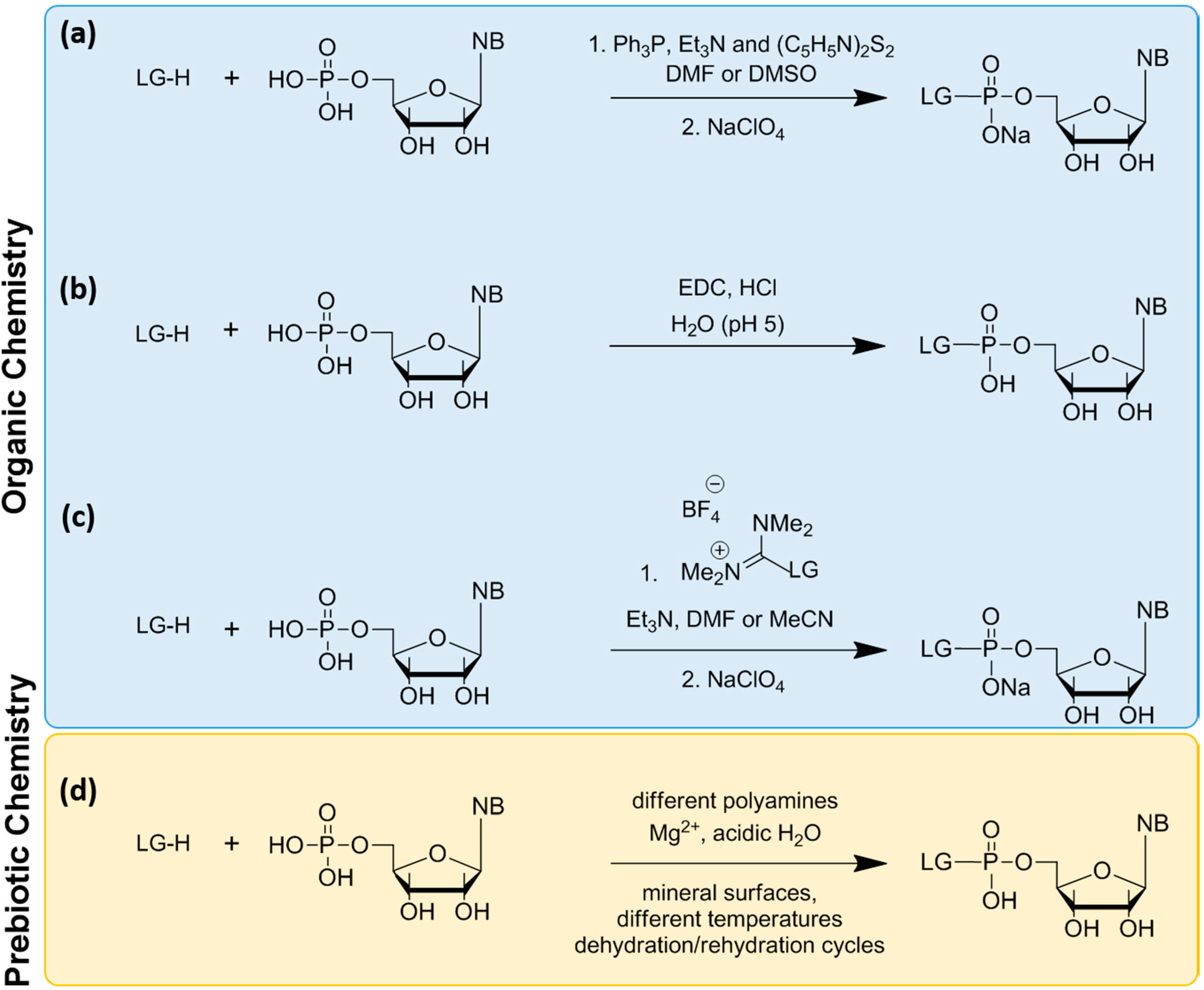
6. Non-Enzymatic RNA Oligomerization
6.1. Mineral-Catalyzed RNA Oligomerization
6.1.1. Effect of Salts, Temperature and pH
6.1.2. Mechanism of Catalysis by Montmorillonite

6.1.3. Effect of Other Minerals
6.2. Lipid-Catalyzed RNA Oligomerization
6.3. “Click Chemistry-Like” RNA Oligomerization in Water
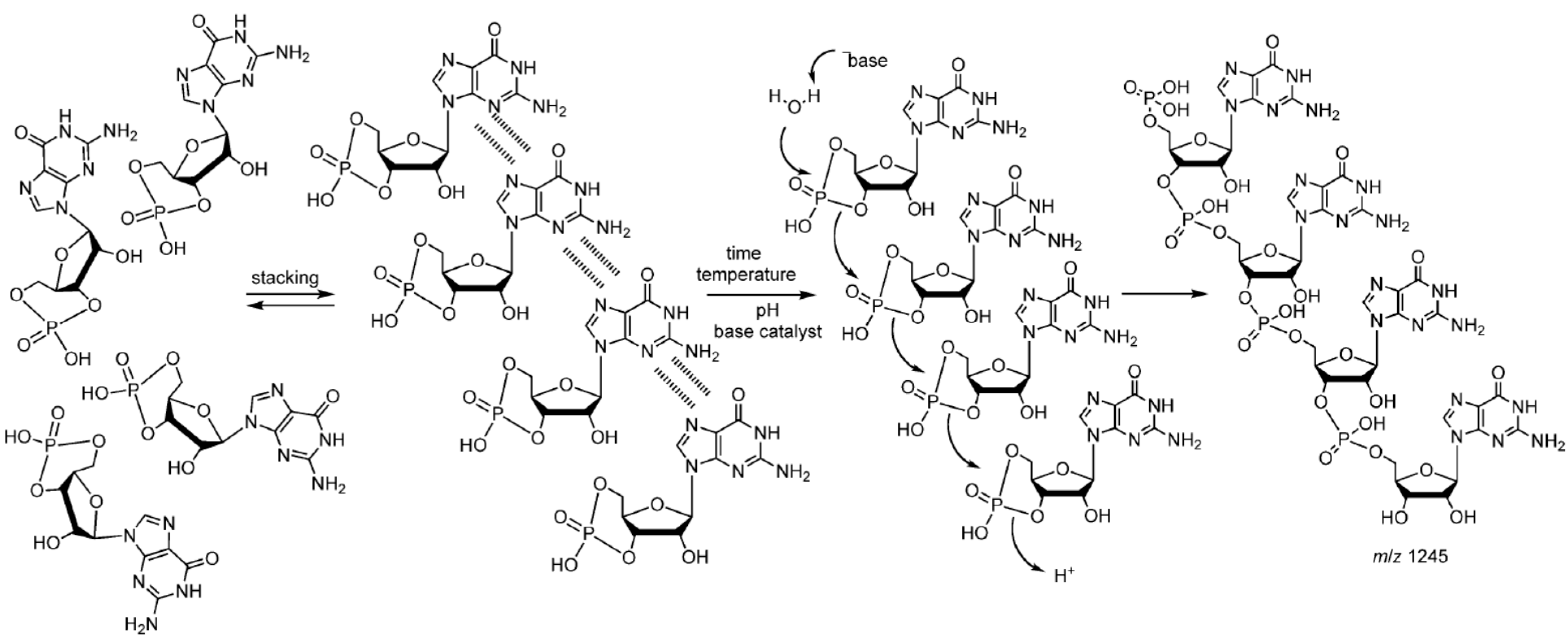

6.4. Peptide-Catalyzed RNA Oligomerization in Eutectic Phase
6.5. Discussion of the Results of Non-Enzymatic RNA Oligomerization
7. A Proposed Experimental Model
8. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Deck, C.; Jauker, M.; Richert, C. Efficient enzyme-free copying of all four nucleobases templated by immobilized RNA. Nat. Chem. 2011, 3, 603–608. [Google Scholar]
- Zhang, S.; Zhang, N.; Blain, J.C.; Szostak, J.W. Synthesis of n3’-p5’-linked phosphoramidate DNA by nonenzymatic template-directed primer extension. J. Am. Chem. Soc. 2013, 135, 924–932. [Google Scholar]
- Adamala, K.; Szostak, J.W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science 2013, 342, 1098–1100. [Google Scholar]
- Schopf, J.W. The first billion years: When did life emerge? Elements 2006, 2, 229–233. [Google Scholar]
- Schidlowski, M. Carbon isotopes as biogeochemical recorders of life over 3.8 Ga of earth history: Evolution of a concept. Precambrian Res. 2001, 106, 117–134. [Google Scholar] [CrossRef]
- Hoffman, P.F.; Schrag, D.P. Snowball earth. Sci. Am. 2000, 282, 68–75. [Google Scholar] [CrossRef]
- Martin, W.; Baross, J.; Kelley, D.; Russell, M.J. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 2008, 6, 805–814. [Google Scholar]
- Lowe, D.R.; Tice, M.M. Tectonic controls on atmospheric, climatic, and biological evolution 3.5–2.4 Ga. Precambrian Res. 2007, 158, 177–197. [Google Scholar] [CrossRef]
- Schwartzman, D.; Lineweaver, C. The hyperthermophilic origin of life revisited. Biochem. Soc. Trans. 2004, 32, 168–171. [Google Scholar] [CrossRef]
- Kasting, J.F. When methane made climate. Sci. Am. 2004, 290, 78–85. [Google Scholar] [CrossRef]
- Newman, M.J.; Rood, R.T. Implications of solar evolution for the earth’s early atmosphere. Science 1977, 198, 1035–1037. [Google Scholar] [CrossRef]
- Bada, J.; Bigham, C.; Miller, S. Impact melting of frozen oceans on the early earth: Implications for the origin of life. Proc. Natl. Acad. Sci. USA 1994, 91, 1248–1250. [Google Scholar] [CrossRef]
- Sleep, N.H. The hadean-archaean environment. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Wilde, S.A.; Valley, J.W.; Peck, W.H.; Graham, C.M. Evidence from detrital zircons for the existence of continental crust and oceans on the earth 4.4 gyr ago. Nature 2001, 409, 175–178. [Google Scholar] [CrossRef]
- Ohmoto, H.; Watanabe, Y.; Ikemi, H.; Poulson, S.R.; Taylor, B.E. Sulphur isotope evidence for an oxic archaean atmosphere. Nature 2006, 442, 908–911. [Google Scholar] [CrossRef]
- Kump, L.R. The rise of atmospheric oxygen. Nature 2008, 451, 277–278. [Google Scholar] [CrossRef]
- Crowe, S.A.; Døssing, L.N.; Beukes, N.J.; Bau, M.; Kruger, S.J.; Frei, R.; Canfield, D.E. Atmospheric oxygenation three billion years ago. Nature 2013, 501, 535–538. [Google Scholar] [CrossRef]
- Valley, J.W. Early earth. Elements 2006, 2, 201–204. [Google Scholar] [CrossRef]
- Ferris, J.P.; Ertem, G. Oligomerization of ribonucleotides on montmorillonite: Reaction of the 5’-phosphorimidazolide of adenosine. Science 1992, 257, 1387–1389. [Google Scholar] [CrossRef]
- Bujdák, J.; Le Son, H.; Yongyai, Y.; Rode, B.M. The effect of reaction conditions on montmorillonite-catalysed peptide formation. Catal. Lett. 1996, 37, 267–272. [Google Scholar] [CrossRef]
- Hanczyc, M.M.; Fujikawa, S.M.; Szostak, J.W. Experimental models of primitive cellular compartments: Encapsulation, growth, and division. Science 2003, 302, 618–622. [Google Scholar] [CrossRef]
- Powner, M.W.; Gerland, B.; Sutherland, J.D. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 2009, 459, 239–242. [Google Scholar] [CrossRef]
- Rajamani, S.; Vlassov, A.; Benner, S.; Coombs, A.; Olasagasti, F.; Deamer, D. Lipid-assisted synthesis of RNA-like polymers from mononucleotides. Orig. Life Evol. Biosph. 2008, 38, 57–74. [Google Scholar] [CrossRef]
- Wieczorek, R.; Dörr, M.; Chotera, A.; Luisi, P.L.; Monnard, P.A. Formation of RNA phosphodiester bond by histidine-containing dipeptides. ChemBioChem 2013, 14, 217–223. [Google Scholar] [CrossRef] [Green Version]
- Hagenbuch, P.; Kervio, E.; Hochgesand, A.; Plutowski, U.; Richert, C. Chemical primer extension: Efficiently determining single nucleotides in DNA. Angew. Chem. Int. Ed. 2005, 44, 6588–6592. [Google Scholar] [CrossRef]
- Vogel, S.R.; Richert, C. Adenosine residues in the template do not block spontaneous replication steps of RNA. Chem. Commun. 2007, 1896–1898. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319. [Google Scholar] [CrossRef]
- Epstein, L.M.; Gall, J.G. Self-cleaving transcripts of satellite DNA from the newt. Cell 1987, 48, 535–543. [Google Scholar] [CrossRef]
- Daròs, J.A.; Flores, R. Identification of a retroviroid-like element from plants. Proc. Natl. Acad. Sci. USA 1995, 92, 6856–6860. [Google Scholar] [CrossRef]
- Ferbeyre, G.; Smith, J.M.; Cedergren, R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell. Biol. 1998, 18, 3880–3888. [Google Scholar]
- Rojas, A.A.; Vazquez-Tello, A.; Ferbeyre, G.; Venanzetti, F.; Bachmann, L.; Paquin, B.; Sbordoni, V.; Cedergren, R. Hammerhead-mediated processing of satellite pDo500 family transcripts from dolichopoda cave crickets. Nucleic Acids Res. 2000, 28, 4037–4043. [Google Scholar] [CrossRef]
- Teixeira, A.; Tahiri-Alaoui, A.; West, S.; Thomas, B.; Ramadass, A.; Martianov, I.; Dye, M.; James, W.; Proudfoot, N.J.; Akoulitchev, A. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature 2004, 432, 526–530. [Google Scholar] [CrossRef]
- Salehi-Ashtiani, K.; Lupták, A.; Litovchick, A.; Szostak, J.W. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 2006, 313, 1788–1792. [Google Scholar] [CrossRef]
- Martick, M.; Horan, L.H.; Noller, H.F.; Scott, W.G. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 2008, 454, 899–902. [Google Scholar] [CrossRef]
- Jimenez, R.M.; Delwart, E.; Lupták, A. Structure-based search reveals hammerhead ribozymes in the human microbiome. J. Biol. Chem. 2011, 286, 7737–7743. [Google Scholar] [CrossRef]
- Przybilski, R.; Gräf, S.; Lescoute, A.; Nellen, W.; Westhof, E.; Steger, G.; Hammann, C. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell Online 2005, 17, 1877–1885. [Google Scholar] [CrossRef]
- De la Peña, M.; García-Robles, I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA 2010, 16, 1943–1950. [Google Scholar] [CrossRef]
- Diener, T. Circular RNAs: Relics of precellular evolution? Proc. Natl. Acad. Sci. USA 1989, 86, 9370–9374. [Google Scholar] [CrossRef]
- Mironov, A.S.; Gusarov, I.; Rafikov, R.; Lopez, L.E.; Shatalin, K.; Kreneva, R.A.; Perumov, D.A.; Nudler, E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 2002, 111, 747–756. [Google Scholar] [CrossRef]
- Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002, 419, 952–956. [Google Scholar] [CrossRef]
- Nahvi, A.; Sudarsan, N.; Ebert, M.S.; Zou, X.; Brown, K.L.; Breaker, R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002, 9, 1043–1049. [Google Scholar] [CrossRef]
- Serganov, A.; Nudler, E. A decade of riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef]
- Steitz, T.A.; Moore, P.B. RNA, the first macromolecular catalyst: The ribosome is a ribozyme. Trends Biochem. Sci. 2003, 28, 411–418. [Google Scholar] [CrossRef]
- Ban, N.; Nissen, P.; Hansen, J.; Moore, P.B.; Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef]
- Nissen, P.; Hansen, J.; Ban, N.; Moore, P.B.; Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science 2000, 289, 920–930. [Google Scholar] [CrossRef]
- Yusupov, M.M.; Yusupova, G.Z.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.H.D.; Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Orgel, L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 99–123. [Google Scholar] [CrossRef]
- Joyce, G.F.; Orgel, L.E. 1 prospects for understanding the origin of the RNA world. Cold Spring Harb. Monogr. Arch. 1993, 24, 1–25. [Google Scholar]
- Maurel, M.C.; Haenni, A.L. The RNA World: Hypotheses, Facts and Experimental Results. In Lectures in Astrobiology; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–594. [Google Scholar]
- Robertson, M.P.; Joyce, G.F. The origins of the RNA world. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef]
- Carter, C.W.; Kraut, J. A proposed model for interaction of polypeptides with RNA. Proc. Natl. Acad. Sci. USA 1974, 71, 283–287. [Google Scholar] [CrossRef]
- Li, L.; Francklyn, C.; Carter, C.W. Aminoacylating urzymes challenge the RNA world hypothesis. J. Biol. Chem. 2013, 288, 26856–26863. [Google Scholar] [CrossRef]
- Engelhart, A.E.; Hud, N.V. Primitive genetic polymers. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Weimann, B.; Lohrmann, R.; Orgel, L.; Schneider-Bernloehr, H.; Sulston, J. Template-directed synthesis with adenosine-5’-phosphorimidazolide. Science 1968, 161. [Google Scholar] [CrossRef]
- Orgel, L.E.; Lohrmann, R. Prebiotic chemistry and nucleic acid replication. Acc. Chem. Res. 1974, 7, 368–377. [Google Scholar] [CrossRef]
- Lohrmann, R. Formation of nucleoside 5’-phosphoramidates under potentially prebiological conditions. J. Mol. Evol. 1977, 10, 137–154. [Google Scholar] [CrossRef]
- Gibbs, D.; Lohrmann, R.; Orgel, L. Template-directed synthesis and selective adsorption of oligoadenylates on hydroxyapatite. J. Mol. Evol. 1980, 15, 347–354. [Google Scholar] [CrossRef]
- Lohrmann, R.; Bridson, P.; Orgel, L. Efficient metal-ion catalyzed template-directed oligonucleotide synthesis. Science 1980, 208, 1464–1465. [Google Scholar] [CrossRef]
- Inoue, T.; Orgel, L. A nonenzymatic RNA polymerase model. Science 1983, 219, 859–862. [Google Scholar] [CrossRef]
- Joyce, G.; Inoue, T.; Orgel, L. Non-enzymatic template-directed synthesis on RNA random copolymers: Poly (C, U) templates. J. Mol. Biol. 1984, 176, 279–306. [Google Scholar] [CrossRef]
- Oró, J.; Basile, B.; Cortes, S.; Shen, C.; Yamrom, T. The prebiotic synthesis and catalytic role of imidazoles and other condensing agents. Orig. Life Evol. Biosph. 1984, 14, 237–242. [Google Scholar] [CrossRef]
- Lohrmann, R.; Orgel, L. Prebiotic activation processes. Nature 1973, 244, 418–420. [Google Scholar] [CrossRef]
- Mukaiyama, T.; Hashimoto, M. Synthesis of oligothymidylates and nucleoside cyclic phosphates by oxidation-reduction condensation. J. Am. Chem. Soc. 1972, 94, 8528–8532. [Google Scholar] [CrossRef]
- Lohrmann, R.; Orgel, L. Preferential formation of (2’–5’)-linked internucleotide bonds in non-enzymatic reactions. Tetrahedron 1978, 34, 853–855. [Google Scholar] [CrossRef]
- Inoue, T.; Orgel, L.E. Substituent control of the poly (C)-directed oligomerization of guanosine 5’-phosphoroimidazolide. J. Am. Chem. Soc. 1981, 103, 7666–7667. [Google Scholar] [CrossRef]
- Ivanovskaya, M.; Gottikh, M.; Shabarova, Z. Modification of oligo (poly) nucleotide phosphomonoester groups in aqueous solutions. Nucleosides Nucleotides 1987, 6, 913–934. [Google Scholar] [CrossRef]
- Prabahar, K.J.; Cole, T.D.; Ferris, J.P. Effect of phosphate activating group on oligonucleotide formation on montmorillonite: The regioselective formation of 3’,5’-linked oligoadenylates. J. Am. Chem. Soc. 1994, 116, 10914–10920. [Google Scholar] [CrossRef]
- Prabahar, K.J.; Ferris, J.P. Adenine derivatives as phosphate-activating groups for the regioselective formation of 3’,5’-linked oligoadenylates on montmorillonite: Possible phosphate-activating groups for the prebiotic synthesis of RNA. J. Am. Chem. Soc. 1997, 119, 4330–4337. [Google Scholar] [CrossRef]
- Röthlingshöfer, M.; Richert, C. Chemical primer extension at submillimolar concentration of deoxynucleotides. J. Org. Chem. 2010, 75, 3945–3952. [Google Scholar] [CrossRef]
- Carpino, L.A.; Imazumi, H.; el-Faham, A.; Ferrer, F.J.; Zhang, C.; Lee, Y.; Foxman, B.M.; Henklein, P.; Hanay, C.; Mügge, C. The uronium/guanidinium peptide coupling reagents: Finally the true uronium salts. Angew. Chem. Int. Ed. 2002, 41, 441–445. [Google Scholar] [CrossRef]
- Vogel, S.R.; Deck, C.; Richert, C. Accelerating chemical replication steps of RNA involving activated ribonucleotides and downstream-binding elements. Chem. Commun. 2005, 4922–4924. [Google Scholar]
- Stütz, J.A.R.; Kervio, E.; Deck, C.; Richert, C. Chemical primer extension: Individual steps of spontaneous replication. Chem. Biodivers. 2007, 4, 784–802. [Google Scholar] [CrossRef]
- Sawai, H.; Lohrmann, R.; Orgel, L. Prebiotic peptide-formation in the solid state. J. Mol. Evol. 1975, 6, 165–184. [Google Scholar] [CrossRef]
- Costanzo, G.; Saladino, R.; Crestini, C.; Ciciriello, F.; di Mauro, E. Nucleoside phosphorylation by phosphate minerals. J. Biol. Chem. 2007, 282, 16729–16735. [Google Scholar] [CrossRef]
- Georgelin, T.; Jaber, M.; Onfroy, T.; Hargrove, A.-A.; Costa-Torro, F.; Lambert, J.-F. Inorganic phosphate and nucleotides on silica surface: Condensation, dismutation, and phosphorylation. J. Phys. Chem. C 2013, 117, 12579–12590. [Google Scholar] [CrossRef]
- Powner, M.W.; Sutherland, J.D.; Szostak, J.W. Chemoselective multicomponent one-pot assembly of purine precursors in water. J. Am. Chem. Soc. 2010, 132, 16677–16688. [Google Scholar] [CrossRef]
- Sutherland, J.D. Ribonucleotides. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Bowler, F.R.; Chan, C.K.; Duffy, C.D.; Gerland, B.; Islam, S.; Powner, M.W.; Sutherland, J.D.; Xu, J. Prebiotically plausible oligoribonucleotide ligation facilitated by chemoselective acetylation. Nat. Chem. 2013, 5, 383–389. [Google Scholar] [CrossRef]
- Lohrmann, R.; Orgel, L. Prebiotic synthesis: Phosphorylation in aqueous solution. Science 1968, 161, 64–66. [Google Scholar] [CrossRef]
- Yamagata, Y.; Inoue, H.; Inomata, K. Specific effect of magnesium ion on 2’,3’-cyclic amp synthesis from adenosine and trimeta phosphate in aqueous solution. Orig. Life Evol. Biosph. 1995, 25, 47–52. [Google Scholar] [CrossRef]
- Li, Y.; Breaker, R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2’-hydroxyl group. J. Am. Chem. Soc. 1999, 121, 5364–5372. [Google Scholar] [CrossRef]
- Scott, W.G.; Szöke, A.; Blaustein, J.; O’Rourke, S.M.; Robertson, M.P. RNA catalysis, thermodynamics and the origin of life. Life 2014, 4, 131–141. [Google Scholar] [CrossRef]
- Verlander, M.; Lohrmann, R.; Orgel, L. Catalysts for the self-polymerization of adenosine cyclic 2’,3’-phosphate. J. Mol. Evol. 1973, 2, 303–316. [Google Scholar] [CrossRef]
- Renz, M.; Lohrmann, R.; Orgel, L. Catalysts for the polymerization of adenosine cyclic 2’,3’-phosphate on a poly (U) template. Biochim. Biophys. Acta (BBA) Nucleic Acids Protein Synth. 1971, 240, 463–471. [Google Scholar] [CrossRef]
- Bernal, J. The physical basis of life. Proc. Phys. Soc. Lond. B 1949, 62. [Google Scholar] [CrossRef]
- Bujdák, J.; Eder, A.; Yongyai, Y.; Faybikova, K.; Rode, B.M. Investigation on the mechanism of peptide chain prolongation on montmorillonite. J. Inorg. Biochem. 1996, 61, 69–78. [Google Scholar] [CrossRef]
- Bujdák, J.; Le Son, H.; Rode, B.M. Montmorillonite catalyzed peptide bond formation: The effect of exchangeable cations. J. Inorg. Biochem. 1996, 63, 119–124. [Google Scholar] [CrossRef]
- Hanczyc, M.M.; Mansy, S.S.; Szostak, J.W. Mineral surface directed membrane assembly. Orig. Life Evol. Biosph. 2007, 37, 67–82. [Google Scholar] [CrossRef]
- Ferris, J.P.; Ertem, G. Montmorillonite catalysis of RNA oligomer formation in aqueous solution. A model for the prebiotic formation of RNA. J. Am. Chem. Soc. 1993, 115, 12270–12275. [Google Scholar] [CrossRef]
- Ertem, G.; Ferris, J.P. Synthesis of RNA oligomers on heterogeneous templates. Nature 1996, 379, 238–240. [Google Scholar] [CrossRef]
- Ferris, J.P.; Hill, A.R.; Liu, R.; Orgel, L.E. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 1996, 381, 59–61. [Google Scholar] [CrossRef]
- Ferris, J.P. Montmorillonite catalysis of 30–50 mer oligonucleotides: Laboratory demonstration of potential steps in the origin of the RNA world. Orig. Life Evol. Biosph. 2002, 32, 311–332. [Google Scholar] [CrossRef]
- Huang, W.; Ferris, J.P. One-step, regioselective synthesis of up to 50-mers of RNA oligomers by montmorillonite catalysis. J. Am. Chem. Soc. 2006, 128, 8914–8919. [Google Scholar] [CrossRef]
- Ertem, G.; Ferris, J.P. Sequence-and regio-selectivity in the montmorillonite-catalyzed synthesis of RNA. Orig. Life Evol. Biosph. 2000, 30, 411–422. [Google Scholar] [CrossRef]
- Miyakawa, S.; Ferris, J.P. Sequence-and regioselectivity in the montmorillonite-catalyzed synthesis of RNA. J. Am. Chem. Soc. 2003, 125, 8202–8208. [Google Scholar] [CrossRef]
- Szostak, J.W. The eightfold path to non-enzymatic RNA replication. J. Syst. Chem. 2012, 3. [Google Scholar] [CrossRef]
- Engelhart, A.E.; Powner, M.W.; Szostak, J.W. Functional RNAs exhibit tolerance for non-heritable 2’–5’ versus 3’–5’ backbone heterogeneity. Nat. Chem. 2013, 5, 390–394. [Google Scholar] [CrossRef]
- Ertem, G. Montmorillonite, oligonucleotides, RNA and origin of life. Orig. Life Evol. Biosph. 2004, 34, 549–570. [Google Scholar] [CrossRef]
- Joshi, P.C.; Aldersley, M.F.; Ferris, J.P. Homochiral selectivity in RNA synthesis: Montmorillonite-catalyzed quaternary reactions of d,l-purine with d,l-pyrimidine nucleotides. Orig. Life Evol. Biosph. 2011, 41, 213–236. [Google Scholar] [CrossRef]
- Joshi, P.C.; Aldersley, M.F.; Ferris, J.P. Progress in demonstrating total homochiral selection in montmorillonite-catalyzed RNA synthesis. Biochem. Biophys. Res. Commun. 2011, 413, 594–598. [Google Scholar] [CrossRef]
- Miyakawa, S.; Joshi, P.C.; Gaffey, M.J.; Gonzalez-Toril, E.; Hyland, C.; Ross, T.; Rybij, K.; Ferris, J.P. Studies in the mineral and salt-catalyzed formation of RNA oligomers. Orig. Life Evol. Biosph. 2006, 36, 343–361. [Google Scholar] [CrossRef]
- Joshi, P.C.; Aldersley, M.F. Significance of mineral salts in prebiotic RNA synthesis catalyzed by montmorillonite. J. Mol. Evol. 2013, 76, 371–379. [Google Scholar] [CrossRef]
- Kawamura, K.; Nakahara, N.; Okamoto, F.; Okuda, N. Temperature dependence of the cyclization of guanine and cytosine mix hexanucleotides with water-soluble carbodiimide at 0–75 °C. Viva Orig. 2003, 31, 221–232. [Google Scholar]
- Kawamura, K. Behaviour of RNA under hydrothermal conditions and the origins of life. Int. J. Astrobiol. 2004, 3, 301–309. [Google Scholar] [CrossRef]
- Joshi, P.C.; Aldersley, M.F.; Delano, J.W.; Ferris, J.P. Mechanism of montmorillonite catalysis in the formation of RNA oligomers. J. Am. Chem. Soc. 2009, 131, 13369–13374. [Google Scholar] [CrossRef]
- Aldersley, M.F.; Joshi, P.C. RNA dimer synthesis using montmorillonite as a catalyst: The role of surface layer charge. Appl. Clay Sci. 2013, 83, 77–82. [Google Scholar] [CrossRef]
- Banin, A.; Lawless, J.; Mazzurco, J.; Church, F.; Margulies, L.; Orenberg, J. pH profile of the adsorption of nucleotides onto montmorillonite. Orig. Life Evol. Biosph. 1985, 15, 89–101. [Google Scholar] [CrossRef]
- Aldersley, M.F.; Joshi, P.C.; Price, J.D.; Ferris, J.P. The role of montmorillonite in its catalysis of RNA synthesis. Appl. Clay Sci. 2011, 54, 1–14. [Google Scholar] [CrossRef]
- Ferris, J.P.; Ertem, G. Oligomerization reactions of ribonucleotides: The reaction of the 5'-phosphorimidazolide of nucleosides on montmorillonite and other minerals. Orig. Life Evol. Biosph. 1992, 22, 369–381. [Google Scholar] [CrossRef]
- Feuillie, C.; Daniel, I.; Michot, L.J.; Pedreira-Segade, U. Adsorption of nucleotides onto fe–mg–al rich swelling clays. Geochim. Cosmochim. Acta 2013, 120, 97–108. [Google Scholar] [CrossRef]
- Toppozini, L.; Dies, H.; Deamer, D.W.; Rheinstädter, M.C. Adenosine monophosphate forms ordered arrays in multilamellar lipid matrices: Insights into assembly of nucleic acid for primitive life. PLoS One 2013, 8. [Google Scholar] [CrossRef]
- DeGuzman, V.; Vercoutere, W.; Shenasa, H.; Deamer, D. Generation of oligonucleotides under hydrothermal conditions by non-enzymatic polymerization. J. Mol. Evol. 2014, 78, 251–262. [Google Scholar] [CrossRef]
- Black, R.A.; Blosser, M.C.; Stottrup, B.L.; Tavakley, R.; Deamer, D.W.; Keller, S.L. Nucleobases bind to and stabilize aggregates of a prebiotic amphiphile, providing a viable mechanism for the emergence of protocells. Proc. Natl. Acad. Sci. USA 2013, 110, 13272–13276. [Google Scholar] [CrossRef]
- Costanzo, G.; Pino, S.; Ciciriello, F.; di Mauro, E. Generation of long RNA chains in water. J. Biol. Chem. 2009, 284, 33206–33216. [Google Scholar] [CrossRef]
- Pino, S.; Ciciriello, F.; Costanzo, G.; di Mauro, E. Nonenzymatic RNA ligation in water. J. Biol. Chem. 2008, 283, 36494–36503. [Google Scholar] [CrossRef]
- Pino, S.; Costanzo, G.; Giorgi, A.; di Mauro, E. Sequence complementarity-driven nonenzymatic ligation of RNA. Biochemistry 2011, 50, 2994–3003. [Google Scholar] [CrossRef]
- Pino, S.; Biasiucci, M.; Scardamaglia, M.; Gigli, G.; Betti, M.G.; Mariani, C.; di Mauro, E. Nonenzymatic ligation of an RNA oligonucleotide analyzed by atomic force microscopy. J. Phys. Chem. B 2011, 115, 6296–6303. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.; Sharpless, K.B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Costanzo, G.; Saladino, R.; Botta, G.; Giorgi, A.; Scipioni, A.; Pino, S.; di Mauro, E. Generation of RNA molecules by a base-catalysed click-like reaction. ChemBioChem 2012, 13, 999–1008. [Google Scholar] [CrossRef]
- Morasch, M.; Mast, C.B.; Langer, J.K.; Schilcher, P.; Braun, D. Dry polymerization of 3’,5’-cyclic gmp to long strands of RNA. ChemBioChem 2014, 15, 879–883. [Google Scholar] [CrossRef]
- Saladino, R.; Crestini, C.; Costanzo, G.; DiMauro, E. On the Prebiotic Synthesis of Nucleobases, Nucleotides, Oligonucleotides, Pre-RNA and Pre-DNA Molecules. In Prebiotic Chemistry; Springer: Berlin/Heidelberg, Germany, 2005; pp. 29–68. [Google Scholar]
- Pino, S.; Costanzo, G.; Giorgi, A.; Šponer, J.; Šponer, J.E.; Mauro, E.D. Ribozyme activity of RNA nonenzymatically polymerized from 3’,5’-cyclic gmp. Entropy 2013, 15, 5362–5383. [Google Scholar] [CrossRef]
- Kanavarioti, A.; Chang, S.; Alberas, D.J. Limiting concentrations of activated mononucleotides necessary for poly (C)-directed elongation of oligoguanylates. J. Mol. Evol. 1990, 31, 462–469. [Google Scholar] [CrossRef]
- Stribling, R.; Miller, S.L. Template-directed synthesis of oligonucleotides under eutectic conditions. J. Mol. Evol. 1991, 32, 289–295. [Google Scholar] [CrossRef]
- Kanavarioti, A.; Monnard, P.-A.; Deamer, D.W. Eutectic phases in ice facilitate nonenzymatic nucleic acid synthesis. Astrobiology 2001, 1, 271–281. [Google Scholar] [CrossRef]
- Bada, J.L.; Lazcano, A. Some like it hot, but not the first biomolecules. Science 2002, 296, 1982–1983. [Google Scholar] [CrossRef]
- Miyakawa, S.; Cleaves, H.J.; Miller, S.L. The cold origin of life: A. Implications based on the hydrolytic stabilities of hydrogen cyanide and formamide. Orig. Life Evol. Biosph. 2002, 32, 195–208. [Google Scholar] [CrossRef]
- Price, P.B. Microbial life in glacial ice and implications for a cold origin of life. FEMS Microbiol. Ecol. 2007, 59, 217–231. [Google Scholar] [CrossRef]
- Levy, M.; Miller, S.L.; Brinton, K.; Bada, J.L. Prebiotic synthesis of adenine and amino acids under europa-like conditions. Icarus 2000, 145, 609–613. [Google Scholar] [CrossRef]
- Cleaves Ii, H.J.; Nelson, K.E.; Miller, S.L. The prebiotic synthesis of pyrimidines in frozen solution. Naturwissenschaften 2006, 93, 228–231. [Google Scholar] [CrossRef]
- Vlassov, A.V.; Johnston, B.H.; Landweber, L.F.; Kazakov, S.A. Ligation activity of fragmented ribozymes in frozen solution: Implications for the RNA world. Nucleic Acids Res. 2004, 32, 2966–2974. [Google Scholar] [CrossRef]
- Adamala, K.; Anella, F.; Wieczorek, R.; Stano, P.; Chiarabelli, C.; Luisi, P.L. Open questions in origin of life: Experimental studies on the origin of nucleic acids and proteins with specific and functional sequences by a chemical synthetic biology approach. Comput. Struct. Biotechnol. J. 2014, 9, 1–10. [Google Scholar] [CrossRef]
- Kasting, J.F. Earth’s early atmosphere. Science 1993, 259, 920–926. [Google Scholar] [CrossRef]
- Corliss, J.B.; Baross, J.A.; Hoffman, S.E. An hypothesis concerning the relationship between submarine hot springs and the origin of life on earth. Oceanol. Acta. 1981, 4, 59–69. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaddour, H.; Sahai, N. Synergism and Mutualism in Non-Enzymatic RNA Polymerization. Life 2014, 4, 598-620. https://doi.org/10.3390/life4040598
Kaddour H, Sahai N. Synergism and Mutualism in Non-Enzymatic RNA Polymerization. Life. 2014; 4(4):598-620. https://doi.org/10.3390/life4040598
Chicago/Turabian StyleKaddour, Hussein, and Nita Sahai. 2014. "Synergism and Mutualism in Non-Enzymatic RNA Polymerization" Life 4, no. 4: 598-620. https://doi.org/10.3390/life4040598



