Abstract
An immune phage library derived from mice, hyperimmunized with morphine-conjugated BSA, was used to isolate a single-chain Fv (scFv) clone, M86, with binding activity to morphine-conjugated thyroglobulin (morphine-C-Tg) but not to codeine-, cocaine-, or ketamine-conjugated Tg. Surface plasmon resonance analysis using a morphine-C-Tg-coupled CM5 sensor chip showed that the Kd value was 1.26 × 10−8 M. To analyze its binding activity to free morphine and related compounds, we performed a competitive ELISA with M86 and morphine-C-Tg in the absence or presence of varying doses of free morphine and related compounds. IC50 values for opium, morphine, codeine, and heroin were 257 ng/mL, 36.4, 7.3, and 7.4 nM, respectively. Ketamine and cocaine exhibited no competitive binding activity to M86. Thus, we established a phage library-derived scFv, M86, which recognized not only free morphine and codeine as opium components but also heroin. This characteristic of M86 may be useful for developing therapeutic reagents for opiate addiction and as a free morphine-specific antibody probe.
1. Introduction
Morphine is one of the most medicinally important analgesics and narcotics. Structurally, it is classified as an alkaloid because of the presence of nitrogen. Its structure is similar to that of codeine, thebaine, and heroin. An immunoassay to accurately discriminate between these analogous alkaloids would be highly beneficial. A key factor for such an assay is specificity with high sensitivity, which is totally dependent on the antibody employed [1,2]. However, most antibodies against haptens are polyclonal serum antibodies that exhibit significant cross-reactivities with closely related compounds.
After a polyclonal anti-morphine antibody was reported in 1971, a number of studies attempted to establish anti-morphine antibodies with higher specificities [3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22]. However, all of these antibodies have shown cross-reactivity among structurally related compounds, particularly with codeine and heroin. For example, Glasel and Sawada reported that their antibody cross-reacted with codeine [4,5]. Usagawa et al. reported that their antibody exhibited cross-reactivity with nalorphine, morphine-6-glucuronide and naltrexone, but not with codeine [9]. Rahbarizadeh et al. [12] established an antibody that had 10% cross-reactivity with heroin and low cross-reactivity with codeine. Dillon et al. [14] produced scFv antibody to morphine-3-glucuronide using a pre-immunized phage display library with a morphine-3-glucuronide-BSA conjugate. This scFv antibody showed cross reactivity with morphine (23.9%), codeine (28.4%) and 6-monoacetylmorphine (15.7%). In 2002, isolation of an scFv antibody that recognized morphine was reported using a pre-immune phage library [15]. However, its specificity to related compounds has not been characterized. Moghaddam et al. [22] reported human scFv antibodies which recognized 6-monoacetylmorphine with high affinities of 1–3 × 10−7 M but not with free morphine.
To develop antibodies that were more specific to morphine, we tested two strategic methodologies to generate morphine-specific antibodies in this study: (1) immunogen preparation (novel types of morphine derivatives and two different conjugated forms) and (2) antibody preparation technology. To prepare immunogens, protein conjugated morphine-6-hemisuccinate, protein conjugated morphine-3-glucuronid, and protein conjugated N-carboxypropylnormorphine in which the 3- and 6-positions and piperidine ring nitrogen atoms were used for protein conjugation as in previous studies [3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22]. Findlay et al. [21] reported that an antiserum against 6-position conjugates recognized structural changes around the piperidine ring nitrogen atom and an antiserum against piperidine ring nitrogen atom conjugates recognized structural changes around the 3- and 6-positions. Thus, to extend the recognition capability of antibodies, we prepared novel types of morphine derivatives (C conjugates) as immunogens, which were attached to carrier proteins via the 2-position. The second method was antibody preparation technology. In antibody preparation technologies, in addition to hybridoma technology, a single chain Fv-displaying phage library has become available, which may make it possible to generate very highly diverse antibody specificity [23].
In this study, we prepared eight types of carrier protein-conjugated narcotics as immunogens or panning antigens to assure specificity to the morphine portion (Figure 1d). These were N conjugates (conjugated through the piperidine ring nitrogen atom of narcotics; Figure 1) and C conjugates (conjugated through the carbon atom at the 2-position of narcotics; Figure 1). N compounds 4, 5, and 6 were synthesized and their protein-conjugated forms were abbreviated as morphine-N-protein (BSA or Tg), cocaine-N-Tg, and ketamine-N-Tg, respectively. A PharmaLink Immunogen kit was used to synthesize Compound 1, its protein-conjugated form was abbreviated morphine-C’-BSA. C compounds 2 and 3 were synthesized and their protein-conjugated forms were abbreviated morphine-C-protein (BSA or Tg) and codeine-C-Tg, respectively. The difference between the C’ and C conjugate was in the linkage portion: that of C’ conjugate was carbonylaminoethylaminomethyl and that of C conjugate was carbonylmethylaminomethyl. During biopanning for an scFv-displaying phage clone, we used morphine-C-conjugated thyroglobulin (Tg; morphine-C-Tg). As a result, we isolated a single clone (M86) that binds to morphine-C-conjugated BSA (morphine-C-BSA) and morphine-C-Tg but not to related compounds conjugated to BSA or Tg. This indicated morphine-specificity to the hapten moiety conjugated to a protein carrier. The Kd value of M86 was 1.26 × 10−8 M. However, a competitive ELISA with free morphine, opium, codeine, and heroin showed that M86 bound to not only free morphine but also to opium, free codeine, and heroin. IC50 values of M86 for opium, morphine, codeine and heroin was 257 ng/mL, 36.4, 7.3 and 7.4 nM, respectively.
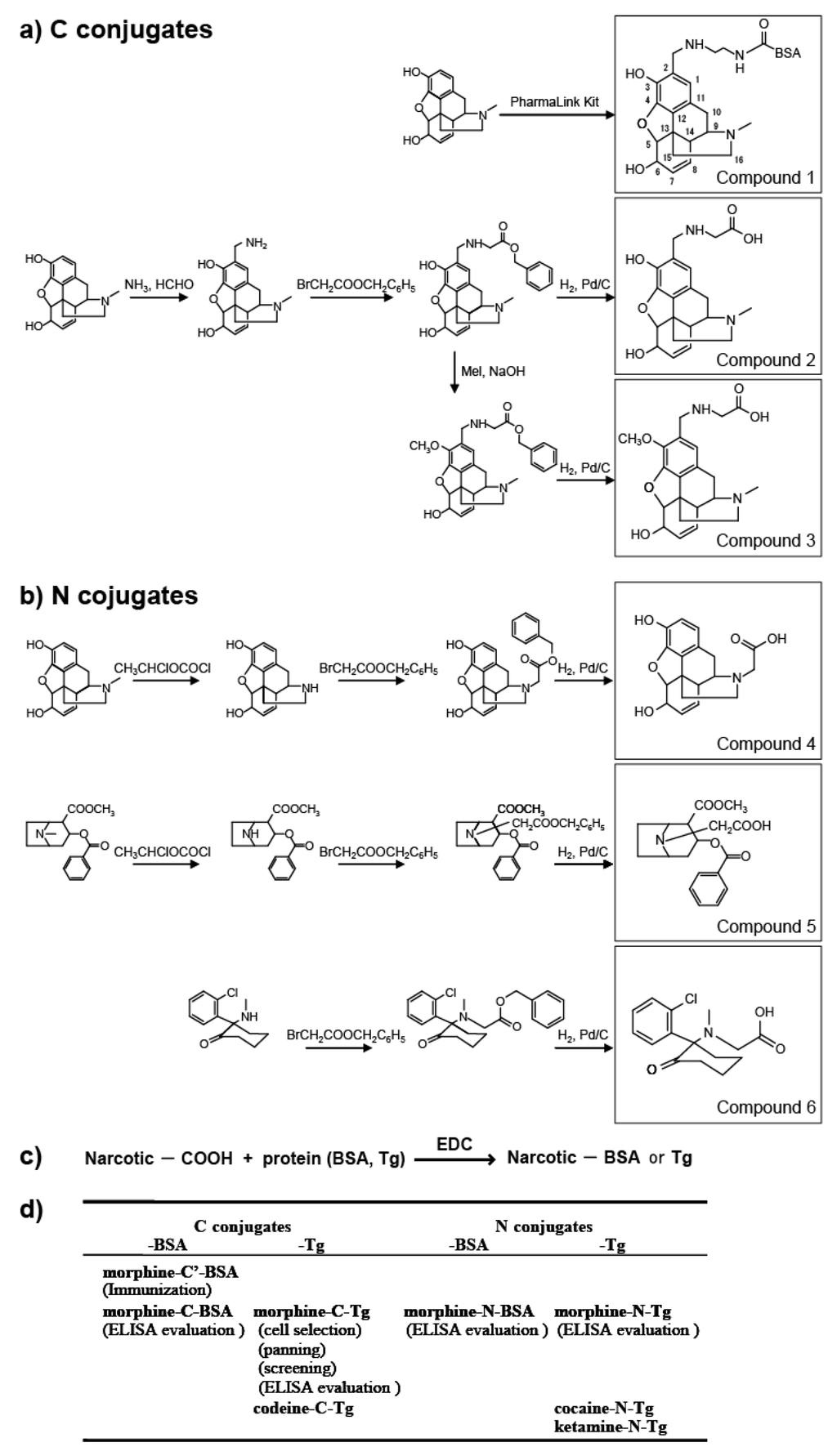
Figure 1.
Synthesis of protein (BSA or Tg) conjugated with narcotics. (a) C conjugate derivatives; (b) N conjugate derivatives; (c) Conjugation of compounds 2–6 with proteins; (d) Summary of antigens used in this study. Parentheses indicate the experimental procedure.
This study presents the results for the development of an scFv antibody, M86, with its detailed characteristics for morphine-binding specificity being defined for the first time. However, this raised a strategic question for using hapten-protein conjugates to establish a free hapten-specific antibody. MOE analysis based on only one X-ray crystallographic structural data of the morphine-reactive antibody 9B1 (PDB ID: IQ0Y, [20]) suggested the possibility of creating a free morphine-specific antibody; therefore, M86 may be useful for developing analytical probes that can specifically detect morphine, codeine, heroin, and therapeutic reagents for opiate addictions.
2. Results and Discussion
2.1. Phage Library Derived from Murine Spleen Cells Immunized with Morphine-Conjugated BSA
The antibody titer against Tg-conjugated morphine was increased in hyperimmunized mice. We confirmed the presence of an antibody that bound strongly to morphine-C-Tg but not to codeine-C-Tg or Tg. Using morphine-C-Tg-coated plastic plates, the binding spleen cells were selected for phage library construction. To prepare phage libraries containing comprehensively diversified V gene repertoires, 128 primer pairs were used to individually amplify the VH or VL genes from the cDNA of these immune splenic cells. Amplified VH and VL products and overlapped scFvs DNA fragments of expected sizes (VH = 386–440 bp; VL = 375–402 bp; and scFv (VH + VL) = 761–842 bp) were obtained. The resulting library included 1.1 × 108 individual clones for the anti-morphine library (Mol-Lib). The quality of Mol-Lib was assessed by ELISA based on E tag detection, which revealed that 70% of the scFv clones in this library had the scFv-g3p fusion protein. DNA sequencing of 63 randomly selected clones revealed that the amino acid sequences of the full-length insert were variable.
2.2. Phage Clones Binding to Morphine-C-Tg
(a) Biopanning. To develop antibodies specific to morphine and with low cross-reactivity with related compounds, we enriched for morphine-specific phage clones using the methodology shown in Figure 1d. Because we confirmed the presence of an antibody in serum that could strongly bind to morphine-C-Tg, but not to codeine-C-Tg or Tg, morphine-C-Tg-coated plastic plates were used to select morphine moiety-reactive lymphocytes for Mol-Lib construction (Figure 1d). Using this phage library, we performed biopanning and screening with morphine-C-Tg (Figure 1d). For ELISA evaluation, to verify the specificity we used both morphine conjugates with different protein carriers and distinct conjugation forms (morphine-N-Tg and morphine-N-BSA; Figure 1d).
Three rounds of phage display were used to pan Mol-Lib in which those that bound to morphine were repeatedly enriched by collecting phages eluted from morphine-C-Tg-coated immunotubes, followed by phage amplification in vivo. A 1000-fold higher enrichment was confirmed during the third round as compared with the first round (Table 1).

Table 1.
Enrichment of the morphine-binding phage.
| Selection round | Input a | Recovery a | Yield b (10−4%) |
|---|---|---|---|
| 1 | 1.2 × 1012 | 6.9 × 103 | 0.0058 |
| 2 | 7.4 × 1011 | 1.6 × 105 | 0.22 |
| 3 | 3.7 × 1011 | 1.7 × 106 | 4.5 |
a Phage inputs and recoveries were determined by infections in E. coli TG1 and plated onto 2TYAG; b Yield was calculated by output phage number/input phage number ×100.
(b) Isolation of morphine-binding phages. A total of 256 clones from the second round of panning and 30 clones from the third round of panning were screened for morphine-binding specificity using morphine-C-Tg. Six morphine-binding clones were obtained: M9, M33, and M38 from the second round of panning and M86, M95, and M101 from the third round of panning (Figure 2a). Binding of this phage clone was detected using an anti-M13 Ab in combination with AP-conjugated goat poly anti-mouse IgG (H + L). DNA sequencing analysis indicated that these six morphine-binding clones were identical, which suggested that specificity to this antigen was rare in the phage library. We characterized clone M86 in this study.
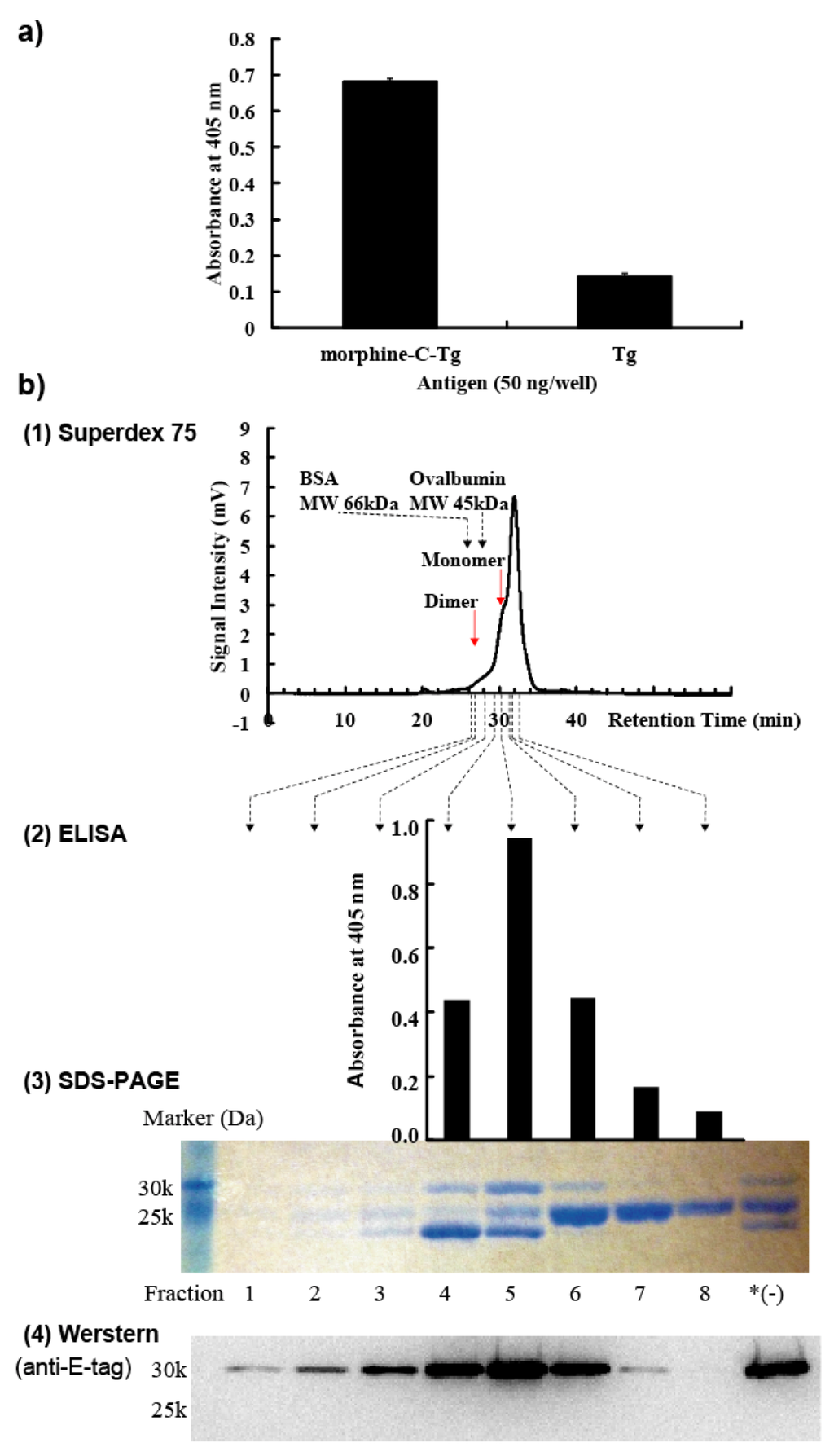
Figure 2.
Biochemical characterization of M86. (a) Phage clone M86 bound to morphine-C-Tg. (b) (1) Size-exclusion HPLC chromatography. Red arrows indicate the positions of the dimer or monomer form of M86. (2) ELISA analysis (upper panel). (3) SDS-PAGE analysis (middle panel). *(−): HisTrap HP column-purified M86. (4) Western blotting analysis (lower panel).
M86 scFv was purified by HisTrap affinity chromatography, followed by Superdex 75 size exclusion chromatography as described in Experimental Section 3.6 (Figure 2b-1). Fractions 1–8 and HisTrap HP column purified but not purified with Superdex 75 were subjected to SDS-PAGE (15%) under reducing conditions (Figure 2b-3) and Western blotting (Figure 2b-4). Fractions 4–8 were added to ELISA plates coated with 50 ng morphine-C-Tg (Figure 2b-2). Detection of Western blotting and ELISA used an anti-E-tag polyclonal antibody in combination with HRP-conjugated goat poly anti-rabbit IgG (H + L). SDS-PAGE analysis resolved 3 bands from these fractions (Figure 2b-3). The binding activity of M86 to morphine-C-Tg was correlated with the appearance of full-length scFv of 30 kDa (Figure 2b-2 and 3). Western blotting analysis revealed that only the 30 kDa band was E-tag-positive (Figure 2b-4). Accordingly, we characterized M86 of 30 kDa for its binding specificity using an anti-E-tag antibody in ELISA analysis.
(c) V gene usage of the M86 gene. The scFv gene sequences of phage clone M86 were examined for CDR usage (Figure 3). CDR1–CDR3 and FR1–FR4 were deduced according to Kabat and Wu [24] (Figure 3a), and germline usage (Figure 3b) was assigned by sequence homology based on the IMGT database for germline V genes. This analysis revealed that M86 was characterized by the germline gene IGHV5-6-4*01, IGHV5-9*02, IGHV5-9-1*01, IGHV5-9-3*01, or IGHV5S9*01 belonging to the VH5 gene family and by IGKV3-4*01 belonging to the Vκ3 gene family.
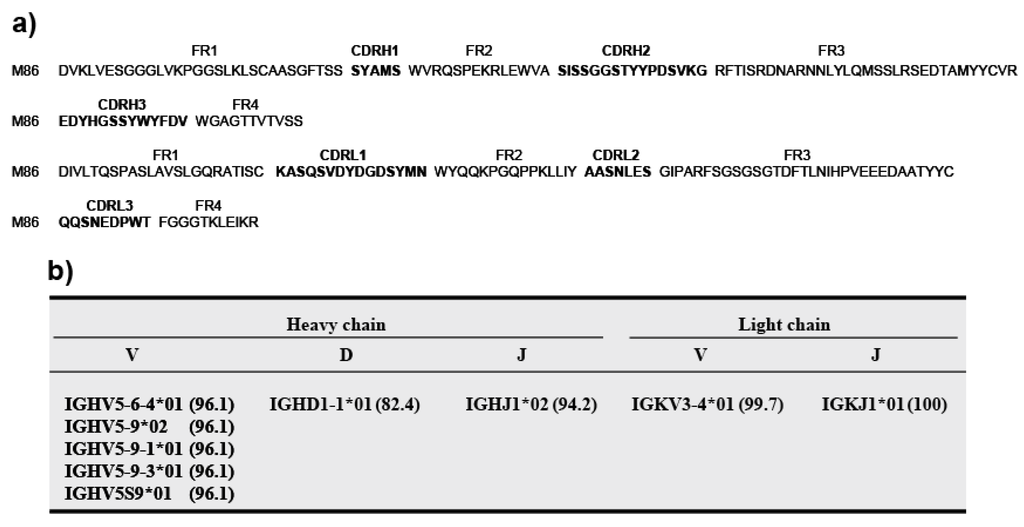
Figure 3.
M86 amino acid sequences and gene usage. (a) Amino acid sequences for the VH and VL domains. DNA sequences were translated to amino acid sequences using Genetyx ver. 8. CDR1–CDR3 and FR1–FR4 were deduced according to Kabat and Wu [24]. CDR regions are indicated in bold. (b) Gene usage. Germline gene usage was assigned based on homology to the IMGT database for germline V genes. Numbers in parentheses indicate the percent identities between the M86 gene and genomic genes.
2.3. M86 Specifically Binds to the Morphine Moieties but not to the Conjugated Portions of Morphine-Conjugated Proteins
We evaluated the binding specificity of M86 to proteins conjugated with morphine-related compounds by ELISA. M86 was incubated in ELISA plates coated with the indicated protein conjugates (Figure 4). M86 binding was detected using an anti-E-tag polyclonal antibody in combination with HRP-conjugated goat poly anti-rabbit IgG (H + L). M86 bound to morphine-C-BSA and morphine-C-Tg but not to BSA or Tg (Figure 4a). M86 bound to morphine-C-Tg in a concentration-dependent manner, but not to codeine-C-Tg (Figure 4b), which indicated that M86 could specifically recognize the morphine moiety but not the conjugation portion. M86 exhibited no binding activity to ketamine-N-Tg or cocaine-N-Tg. These results suggested that M86 recognized the neighborhood of the hydroxyl group moiety at the 3-position of morphine as long as the morphine moiety was conjugated to a protein carrier (Figure 4b).
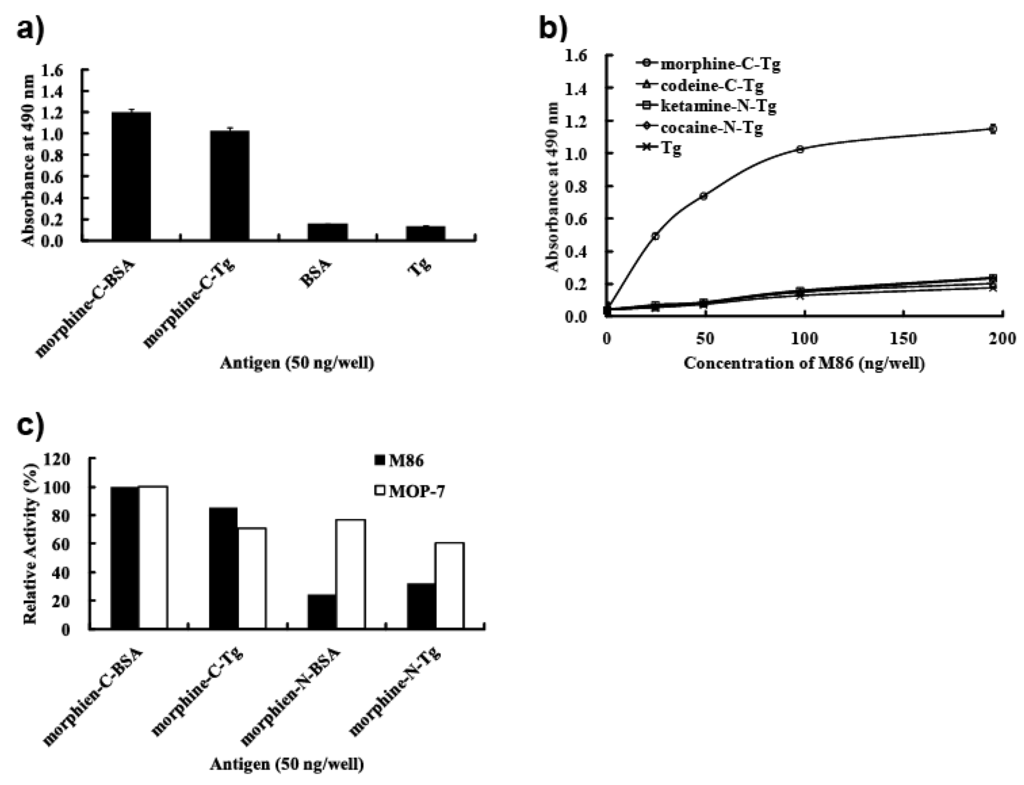
Figure 4.
M86 binding specificity. (a) M86 specifically binds to the morphine moiety but not to the carrier protein. (b) Dose response of M86 for binding to morphine-C-Tg, codeine-C-Tg, and unrelated narcotic-conjugated proteins. (c) Preferential binding activity of M86 to C conjugates compared with N conjugates.
We evaluated the reactivity of M86 to the morphine moiety by using C conjugates (morphine-C-BSA and morphine-C-Tg) and N conjugates (morphine-N-BSA and morphine-N-Tg) (Figure 4c). Their binding activities are represented as relative activities (%) at A490 compared with those against morphine-C-BSA (A490 = 1.199 for M86 and 0.602 for MOP 7). M86 showed higher reactivity with C conjugates in contrast to its relatively low reactivity with N conjugates, whereas a commercial monoclonal anti-morphine antibody (MOP-7) showed similar reactivity levels with these C and N conjugates (Figure 4c). These results suggested that M86 recognized not only the neighborhood of the hydroxyl group at the 3-position but also the neighborhood of the nitrogen atom.
2.4. SPR Analysis
For this analysis, a monomeric M86 scFv fraction was prepared with Superdex 75, which confirmed the monomer form of M86 in fraction 5 (Figure S1). To determine the molecular species with morphine-C-Tg-binding activity, the affinity-purified M86 with a HisTrap HP column was incubated in immunotubes coated with various proteins including morphine-C-Tg and recoverd by eluting with glycine–HCl (pH 2.2) (Experimental Section 3.5). SDS-PAGE analysis using these elutes demonstrated that two bands from the top but not the bottom band of the gel bore the binding activity to morphine-C-Tg (Figure 5). These two bands showed no binding activity to Tg, ovalbumin, or ketamine-N-Tg. Therefore the 2nd band from the top may be a truncated product of M86 where the E-tag is cleaved off. Although the structure of the 2nd band remains to be solved, the protein concentration of these two bands were estimated by densitometric analysis (ImageJ) of SDS-PAGE. SPR analysis was performed using CM5 sensor chips conjugated with morphine-C-Tg. Based on the data obtained, the Kd of M86 was estimated to be 1.26 × 10−8 M (Figure 6).
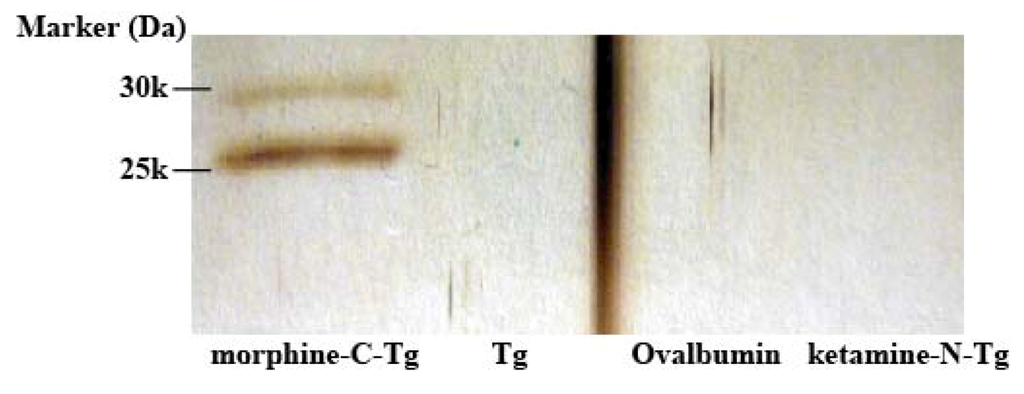
Figure 5.
Morphine-binding activity of affinity-purified M86. Affinity-purified M86 with HisTrap HP column (1 µg/0.5 mL) was incubated in an immunotube coated with morphine-C-Tg, Tg, Ovalbumin, or ketamine-N-Tg (2 µg/0.5 mL) as described in Experimental Section 3.5. The bound M86 was eluted with 0.1 M glycine–HCl (pH 2.2). The eluted M86 was subjected to 15% SDS-PAGE under reducing conditions and was visualized using silver staining.
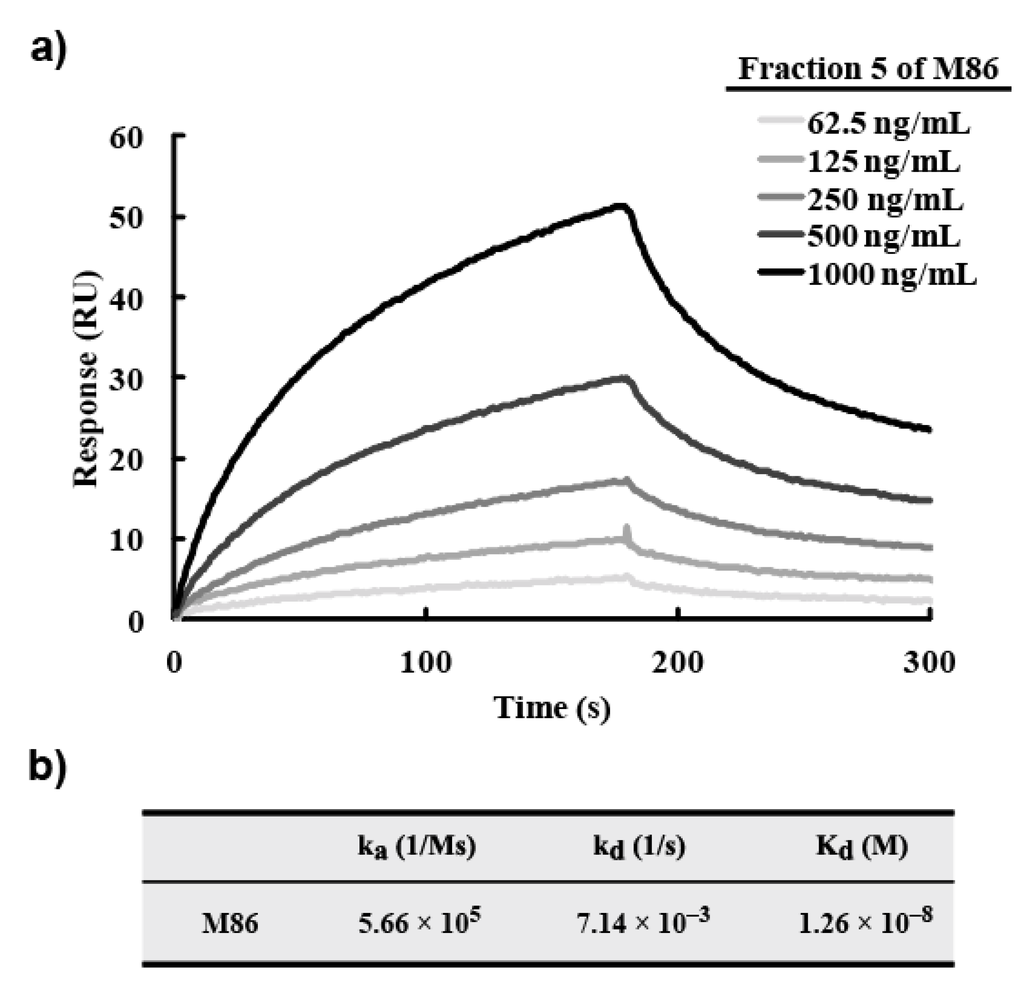
Figure 6.
SPR analysis for M86. (a) SPR sensorgram recordings for the association and dissociation of M86. Morphine-C-Tg (20 µg/mL) was immobilized on CM5 sensor chips as described in Experimental Section 3.10. The protein concentration of fraction 5 was indicated. (b) Summary. The protein concentration of morphine-C-Tg-binding M86 (1st and 2nd bands of fraction 5: see Figure 2b-3) was estimated to be 59% by densitometric analysis (ImageJ) and used to estimate the Kd value. Kd was calculated according to 1:1 binding model.
2.5. M86 Binds not only to Free Morphine but also to Codeine and Heroin
To analyze the binding activity of M86 to free morphine and related compounds (codeine, heroin, or opium), we performed a competitive ELISA for M86 and morphine-C-Tg in the absence or presence of varying doses of free morphine, related compounds, or unrelated compounds (Figure 7). M86 exhibited binding activity not only to free morphine but also opium, free codeine, and heroin (Figure 7a). Employing GraphPad Prism software, IC50 values were 36.4 nM (morphine), 257 ng/mL (opium), 7.3 nM (codeine), and 7.4 nM (heroin) (Figure 7). In contrast, M86 did not show binding activity to cocaine, ketamine (Figure 7a), or Tg (Figure 7b). Its reactivity with codeine and heroin were higher than that with morphine. The Ki values were also estimated to be 18.9 nM (morphine), 4.0 nM (codeine), and 5.2 nM (heroin).
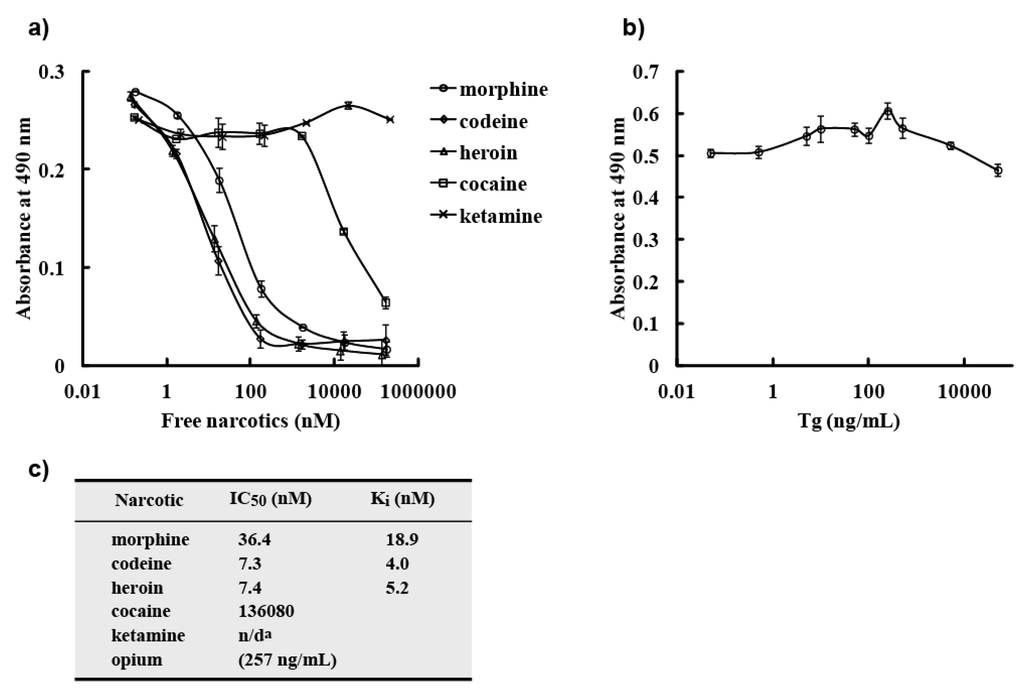
Figure 7.
Competitive ELISA. (a) M86 binding to morphine-C-Tg was competitively inhibited by varying doses of free morphine or related compounds in ELISA. M86 (75 ng) was incubated with varying concentrations of free morphine, codeine, heroin, opium, cocaine, or ketamine and added to ELISA plates coated with 50 ng of morphine-C-Tg. M86 binding was detected using an anti-E-tag polyclonal antibody in combination with HRP-conjugated goat poly anti-rabbit IgG (H + L). All experiments were done in triplicate. (b) M86 binding to morphine-C-Tg was not competitively inhibited by varying doses of Tg in ELISA. The experiments were performed as described in (a). (c) Summary of IC50 values. IC50 and Ki values were determined using GraphPad Prism software. n/da: could not be determined.
2.6. Computer Simulations of M86
When the binding specificity of M86 to hapten-conjugated proteins was analyzed by ELISA, M86 recognized the morphine moiety. Despite our methodological interventions, competitive ELISA showed that M86 could not discriminate free codeine or heroin from morphine. Thus, this study suggested a methodological difficulty for isolating free hapten-specific antibodies because both the immunization procedures and screening system involved hapten-conjugated proteins. Thus, we tested the usefulness of computer simulations to characterize the binding characteristics of M86, because X-ray crystallographic structural data of the anti-morphine antibody 9B1 (PDB ID: 1Q0Y, [20]) was available, although the cross-reactivity of this antibody was not characterized. X-ray crystallographic structural data for anti-morphine antibody 9B1 (PDB ID: 1Q0Y) was used for MOE analysis. The ligand interaction surface of 9B1 is highlighted in the left panel (Figure 8a). 9B1 is depicted as a ribbon structure in gray, and the oriented morphine is shown by a green stick model. The ligand interaction surface of M86 is highlighted in the right panel (Figure 8a). M86 is depicted as a ribbon structure in yellow, and the oriented morphine is shown using a blue stick model. The amino acid residues around the ligand interaction site are shown as stick models using the numbering of Kabat and Wu [24]. The green arrow indicates the C-conjugation site of morphine with protein, and the orange arrow indicates the N-conjugation site of morphine with protein. 9B1 bound to the cationic nitrogen atom of morphine and the polar oxygen atoms (3-, 6-position) of morphine exposed to solvent [20,25], while M86 appeared to bind to the polar oxygen atoms (3-, 6-position) of morphine (Figure 8b). This suggested that the antigen recognition modes of these antibodies were not identical. However, because 9B1 data were only for morphine-reactive antibody, we performed computer simulation analysis of M86 on the basis of 9B1 data. The 9B1 amino acid sequence was presented in comparison with that of M86 (Figure 8c). Homology analysis was performed by Genetyx ver. 8.
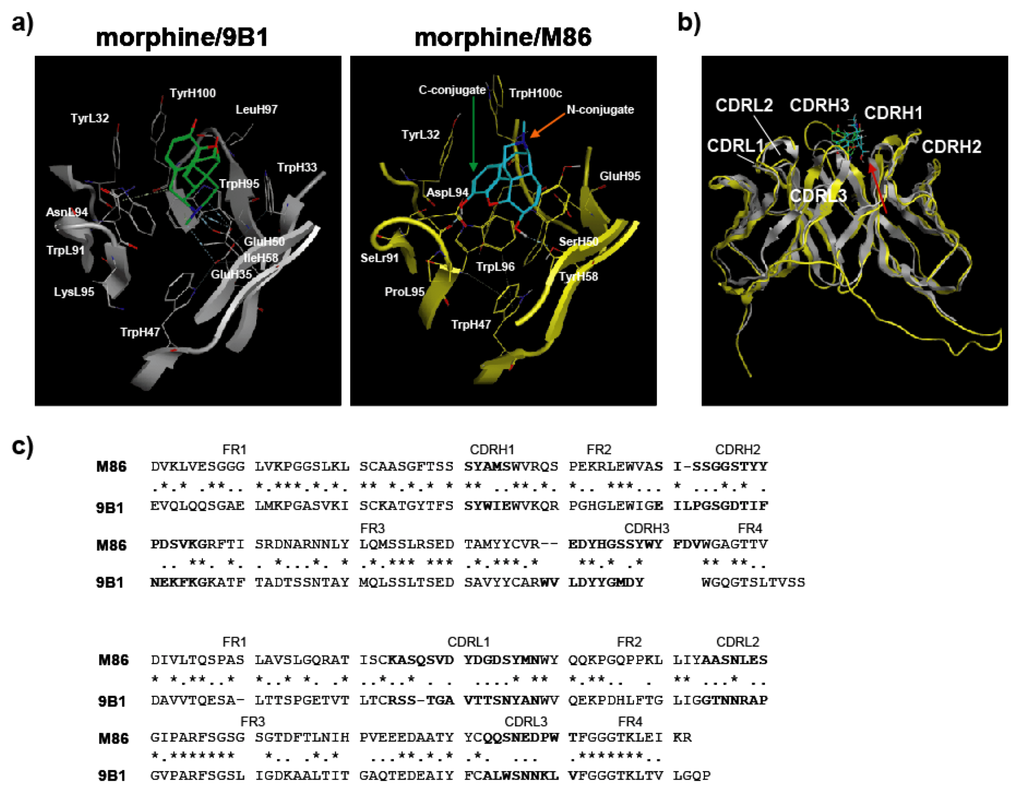
Figure 8.
Computer simulations for the structure of M86 scFv. (a) Side view of the ligand interaction surface of 9B1 and M86. (b) Superposition of M86 with 9B1. The red arrow indicates the direction of the side view in (a). (c) Amino acid sequences of 9B1 and M86. Asterisks and dots indicate identical residues and similar residues, respectively.
This analysis indicates that SerL91 and AspL94 of M86 are located near the 3-hydroxyl group (methoxy group for codeine and methoxycarbonyl group for heroin), and these amino acid substitutions for those that bear bulky side functional groups may selectively inhibit the binding activity to codeine or heroin while maintaining the binding activity to morphine (Figure 9). This suggested that a computer simulation was a powerful approach for developing an antibody with high specificity, particularly for a free hapten-specific antibody.
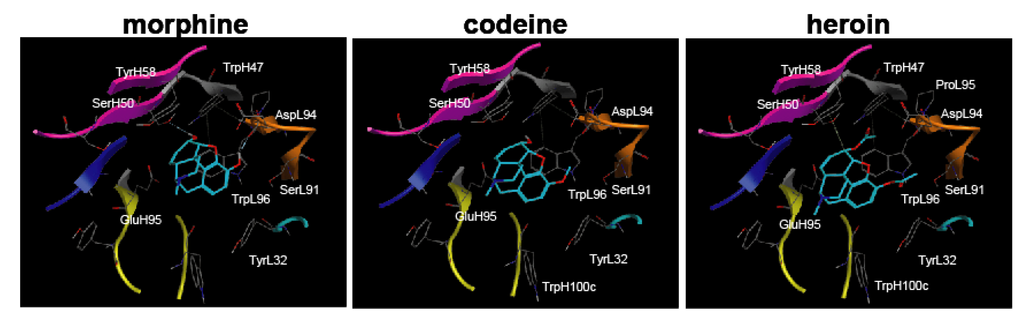
Figure 9.
Binding models of morphine-related compounds to M86 scFv indicated by a top view of the M86 binding pocket. The structure of M86 scFv was simulated as described in Figure 7. M86 is shown as a ribbon structure (CDRH1, cyan; CDRH2, magenta; CDRH3, yellow; CDRL1, blue; CDRL2, red; CDRL3, orange), and narcotics are shown as blue stick structures for morphine (left), codeine (middle), and heroin (right). The amino acid residues around the ligand interaction site are shown as a stick model using the numbering of Kabat and Wu [24]. The hydrogen bond interaction is denoted by a dotted line. Left panel: M86 binding to morphine; Middle panel: M86 binding to codeine; Right panel: M86 binding to heroin.
3. Experimental Section
3.1. Antigen Synthesis
We prepared (5α,6α)-7,8-didehydro-4,5-epoxy-17-methylmorphinan-3,6-diol (morphine), (5α,6α)-7,8-didehydro-4,5-epoxy-3-methoxy-17-methylmorphinan-6-ol (codeine), (5α,6α)-7,8-didehydro-4,5-epoxy-17-methylmorphinan-3,6-diol diacetate (heroin), (RS)-2-(2-chlorophenyl)-2-methylamino-cyclohexan-1-one (ketamine), and methyl (1R,2R,3S,5S)-3-(benzoyloxy)-8-methyl-8-azabicyclo[3.2.1]octane-2-carboxylate (cocaine). All narcotics were handled at the Central Customs Laboratory or Kyushu University with permission for their use. Three different conjugated forms of hapten conjugates (C’ conjugate, C conjugate, and N conjugate) were synthesized as described below (Figure 1, compounds 1–6).
3.1.1. C Position-Conjugated Derivatives (Figure 1a, Compounds 1–3)
A Thermo Scientific® PharmaLink Immunogen Kit (77158; Thermo Scientific, Waltham, MA, USA) was used to synthesize compound 1 (morphine-C’-BSA) from morphine by the Mannich reaction according to the manufacturer’s instructions. Morphine (10 µmol) was incubated with SuperCarrier® at room temperature for 24 h. The immunogen was purified by gel filtration. Compounds 2 and 3 were synthesized as described below. After aminomethylation of morphine (30 µmol) by the Mannich reaction, the 2-aminomethylated compound was mixed with bromoacetic acid benzyl ester (30 µmol) to give the benzyloxycarbonylmethylaminomethyl-conjugated intermediate. The benzyloxycarbonyl-methylaminomethyl-conjugated intermediate (ca. 10 µmol) was reduced under hydrogen in the presence of Pd/C to give hydroxycarbonylmethylaminomethyl-conjugated derivative compound 2. The benzyloxycarbonylmethylaminomethyl-conjugated intermediate (ca. 10 µmol) was treated with 10 µmol methyl iodide in the presence of sodium hydride to give the corresponding product, 3-methoxy-2-benzyloxycarbonylmethylaminomethyl (intermediate). This intermediate was reduced in a manner similar to that used for synthesizing compound 2 to give compound 3.
3.1.2. N Position-Conjugated Derivatives (Figure 1b, Compounds 4–6)
1-Chloroethyl chloroformate was used to synthesize normorphine and norcocaine by N-demethylation of morphine and cocaine, respectively [26]. Morphine or cocaine (20 µmol) was mixed with four equivalents of 1-chloroethyl chloroformate, and the mixture was refluxed for 15 h under nitrogen to give normorphine or norcocaine depending on the starting compound. Normorphine and norcocaine (10 µmol) were reacted separately with bromoacetic acid benzyl ester following reduction using Pd/C under hydrogen to give compounds 4 and 5, respectively, in a manner similar to compound 2 synthesis. Ketamine (20 µmol) was mixed with bromoacetic acid benzyl ester (22 µmol) to give the N-benzyloxy-carboxymethyl derivative, which was reduced by hydrogen over a Pd/C catalyst to give compound 6.
3.1.3. Protein Conjugation (Figure 1c)
An aqueous solution of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC) was used to activate the hydroxycarbonyl side chain-linked narcotic (compounds 2–6; ca. 10 µmol) for coupling. The solution was added to 10 mg of thyroglobulin (Tg) in 2 mL of 50 mM phosphate buffer (pH 7.5), stirred at 4 °C for 15 h, and cleaned up by gel filtration. The same protocol was used to couple compounds 2 and 4 to bovine serum albumin (BSA).
3.2. Immunization
We used 7-week-old female BALB/c mice (n = 3; Charles River Laboratories Japan, Inc., Yokohama, Japan). Morphine-C′-BSA with complete Freund’s adjuvant (263810; Difco Laboratories, Detroit, MI, USA) was intraperitoneally administered at a dose of 25 µg/100 µL morphine-C’-BSA/phosphate buffered saline (PBS). Subsequently, each mouse was booster immunized on days 14, 28, 42, and 56 with the same amount of morphine-C′-BSA together with incomplete Freund’s adjuvant (263910; Difco Laboratories, Detroit, MI, USA). Blood was collected through the orbital vein of the mouse eye and its serum antibody titer was determined by ELISA. Three days after the fifth immunization, using spleen cells derived from mice hyperimmunized with morphine-C′-BSA, morphine moiety-reactive lymphocytes were enriched with morphine-C-Tg-coated on plastic plates. All animal experiments were performed under the laboratory guidelines for Animal Experiments of Kagoshima University.
3.3. Single-Chain Library Construction
A phage library was constructed according to Burmester et al. [27]. Total RNA was purified from the morphine moiety-reactive lymphocytes, which were enriched from mice spleen cells using morphine-C-Tg-coated on plastic plates. VH and VL genes were amplified from cDNA that was prepared as previously described [27]. The primers used for amplifying VH and VL genes and overlapping regions were according to the previously described primer set [27] except that the integration order of DNA segments was changed from the original sequence, 5′-SfiI site-VL-linker-VH- SfiI site-3′ to 5′-SfiI site-VH-linker-VL-SpeI site-3′. In addition, no FLAG was used. For VH gene amplification, 76 separate PCR amplifications were performed using 19 different VH forward primers (HF) and four different VH back primers (HB). For Vκ genes, 51 separate PCR amplifications were performed using 17 different Vκ forward primers (LF) and three different Vκ back primers (LB). For Vλ genes, one PCR amplification was performed using a Vλ forward primer (LFλ) and a Vλ back primer (LBλ). The scFv genes were generated by assembly of VH and VL genes through a (Gly4Ser)3 linker sequence by PCR with an scFv forward primer and an scFv back primer. The scFv genes were digested with SfiI (R0123S; New England Biolabs Inc., Ipswich, MA, USA) and SpeI (R0133L; New England Biolabs Inc.), agarose gel purified, and ligated into the phagemid vector pCANTAB/5E. The ligation products were electroporated into Escherichia coli TG1 DUOs Phage Display Electrocompetent Cells (60502-2; Lucigen Corporation, Middleton, WI, USA). Library diversity was defined as the number of recombinant single ampicillin-resistant clones obtained from consecutive ligations and transformations of the plasmid into competent cells prior to any amplification procedures for the genes. Clones were selected at random, and the insert plasmid DNA was sequenced and analyzed with an Applied Biosystems 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
3.4. ScFv-displaying Phage Preparation
ScFv-displaying phage preparation was as described by Hashiguchi et al. [28]. E. coli TG1 cells harboring the constructed phagemids were shaken in a 2YT medium supplemented with 100 μg/mL ampicillin and 2% glucose (2YTAG medium) at 37 °C until the A600 reached approximately 0.5, and the M13KO7 helper phage [final, ca. 109 colony-forming units (cfu)/mL] were added. After 30 min incubation at 37 °C, the culture was shaken for 30 min at 37 °C. After centrifugation, the cells were suspended in 2YT supplemented with 100 μg/mL ampicillin and 25 μg/mL kanamycin. The culture was then shaken overnight at 30 °C. Phage particles were purified by precipitation with 0.2 volume of 12.5% (w/v) polyethylene glycol (PEG) 6000 and 2.48 M sodium chloride. The scFv-displaying phages were resuspended in PBS.
3.5. Biopanning
The library was subjected to three rounds of panning as described by Hashiguchi et al. [28]. An immunotube (Maxisorp; Nunc, Roskilde, Denmark) was coated with 1 µg of morphine-C-Tg. After blocking with 1% BlockAce (UK-B40; DS Pharma Biomedical, Osaka, Japan), 1012 cfu phage particles in 1 ml of 1% BSA plus 1% Tg that were pre-adsorbed to blocking solution were added to the immunotube and incubated for 1 h. The immunotube was washed 10 times with PBS/0.1% Tween 20 (PBST) followed by five times with PBS. Bound phages were eluted by adding 1 ml of 0.1 M glycine–HCl (pH 2.2). Eluted phages were amplified by infection in fresh E. coli TG1 cells, and the phages were rescued as described earlier. To recover morphine-binding component, 1 µg/0.5 mL of affinity-purified M86 with HisTrap HP column was incubated for 2 h in an immunotube coated with 2 µg/0.5 mL of morphine-C-Tg, Tg, Ovalbumin or ketamine-N-Tg. Blocking was performed with 1% BlockAce, and washing was performed 5 times with PBST followed by once with PBS. Elution was in a manner similar to that for phage panning, except that 0.5 mL of 0.1 M glycine–HCl (pH 2.2) was added.
3.6. Soluble ScFv Preparation
Soluble scFv was prepared by infecting phage clones in E. coli HB2151 and then incubating with 1 mM IPTG for 12 h at 25 °C as described by Hashiguchi [28]. Cell pellets were collected by centrifugation, and resuspended in 1 mM EDTA/PBS. After incubation on ice for 30 min, periplasmic fraction was collected by centrifugation. A HisTrap HP column (17-0404-03; GE Healthcare, Princeton, NJ, USA) was used to purify these periplasmic scFvs according to the manufacturer’s recommendations. Eluted fractions were dialyzed against PBS. To prepare the monomeric M86 scFv fraction, affinity-purified M86 with a HisTrap HP column was fractionated to 100 µL by Superdex 75 10/300 GL (17-5174-01; GE Healthcare), which was developed with PBS at a flow rate of 0.4 ml/min. BSA and ovalbumin were used as molecular size markers.
3.7. Western Blotting
Western blotting analysis was performed as described by Hashiguchi [28]. Fractionated scFvs were subjected to 15% SDS-PAGE under reducing conditions and the gel was blotted onto a PVDF membrane (Millipore, MA, USA). Detection was performed using an anti-E-tag polyclonal antibody in combination with HRP-conjugated goat poly anti-rabbit IgG and visualized by Chemi-Lumi One (Nacalai Tesque Inc., Kyoto, Japan) and LAS-1000 (Fuji Film, Tokyo, Japan).
3.8. ELISA and Competitive ELISA
ELISA was performed as described previously [28]. A Nunc-Immuno plate (Maxisorp, Nunc, Roskilde, Denmark) was coated with BSA- (or Tg)-conjugated narcotic antigens (50 ng in 50 µL/well) or control protein. Each well was blocked with 1% BlockAce. For scFv-displayed phage detection, 50 µL of diluted PEG-precipitated phage clones (dilution of 1:50/well) was added and incubated for 2 h. To detect bound phage clones, the clones were incubated with an anti-M13 monoclonal antibody (27-9420-01; GE Healthcare) at a dilution of 1:1,000 as a primary antibody for 1 h and with alkaline phosphatase (AP)-conjugated goat poly anti-mouse IgG (H + L) (115-055-003; Jackson ImmunoResearch, West Grove, PA, USA) at a dilution of 1:1000 as a secondary antibody for 30 min. A microplate reader (EnSpire 2300; PerkinElmer, Waltham, MA, USA) was used to measure absorbance at 405 nm after incubation with 50 µl of a p-nitrophenyl phosphate/10% diethanol amine solution. For soluble scFv detection, diluted scFvs was incubated for 2 h, followed by incubation with an anti-E-tag polyclonal antibody (PC-229; Kamiya Biomedical Company, Seattle, WA, USA) at a dilution of 1:1,000 as a primary antibody for 1 h and horseradish peroxidase (HRP)-conjugated goat poly anti-rabbit IgG (H + L) (31462; Thermo Scientific, Waltham, MA, USA) at a dilution of 1:1,000 as a secondary antibody for 30 min. After incubation with a SigmaFast OPD tablet set (P9187-50SET; Sigma–Aldrich Corporation, Munich, Germany), absorbance was measured at 490 nm. Detection by MOP-7 (murine anti-morphine IgG: sc-69864; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was in a manner similar to that for soluble scFv detection, except that diluted MOP-7 (1:5,000) was added to each well in place of scFv and HRP-conjugated goat poly anti-mouse IgG (sc-2031; Santa Cruz Biotechnology) at a dilution of 1:2,500 was used as a secondary antibody. Competitive ELISA was done in a manner similar to that for soluble scFv detection, except that pre-mixed solutions of scFv and free narcotic were added to each well in place of scFv.
3.9. DNA Sequencing
Applied Biosystems 3130xl Genetic Analyzer and BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) was used to identify the nucleotide sequences of the scFv genes. Genetyx ver. 8. software (Genetyx Corporation, Tokyo, Japan) was used to analyze nucleotide sequences. Amino acid sequences deduced from the nucleotide data were aligned and presented in the format described by Kabat and Wu [24]. The scFv nucleotide sequences were analyzed by searching the IMGT/V-QUEST database. The complementary determining regions (CDR1–CDR3) and flanking regions (FR1–FR4) were deduced according to Kabat and Wu [24].
3.10. Surface Plasmon Resonance Analysis
A BIAcore X100 system (GE Healthcare) was used according to the manufacturer’s instructions. Morphine-C-Tg (20 µg/mL of 10 mM sodium acetate buffer, pH 4.0) was immobilized onto a CM5 sensor chip according to the amine-coupling protocol provided by the manufacturer, and unreacted sites were masked with 1 M ethanolamine–HCl (pH 8.5). The association reaction was initiated by injecting varying concentrations of scFv solutions. Analyte injection was at a flow rate of 30 µL/min. The dissociation reaction was begun by washing with HBS–EP buffer (0.01 M Hepes, 0.15 M NaCl, 3 mM EDTA, 0.005% v/v surfactant P20, pH 7.4). At the end of a cycle, the sensor chip surface was regenerated with a 0.1 M glycine–HCl buffer (pH 2.5). BIAcore system software (BIAcore X100 evaluation software) was used to calculate the association (ka, M−1s−1) and dissociation (kd, s−1) constants. Kd (M) was calculated according to the 1:1 binding model.
3.11. Computer Simulations
Molecular Operating Environment (MOE) software (Chemical Computing Group Inc., Montreal, Canada) [29] was used for all molecular modeling studies. The X-ray crystallographic structure of the anti-morphine antibody 9B1 (PDB ID: 1Q0Y) [20] was obtained from a protein data bank as a template for homology modeling of M86. The MMFF94s force field and the generalized Born salvation model were used for the minimization step. After homology modeling, M86 was prepared for docking studies in which (i) hydrogen atoms were added to the structure with standard geometry; (ii) the structure was minimized using a MMFF94s force field; (iii) MOE Alpha Site Finder was used for active site searches within M86, and dummy atoms were created from the obtained alpha spheres; and (iv) the obtained M86 structure was then used in the ASEDock program (Ryoka Systems Inc., Tokyo, Japan) [30] with a narcotics library in which the conformational analysis data for morphine, codeine, and heroin were determined by the MOE program. The complexes were evaluated by Udock scores, which provided the affinities between the ligands and M86.
4. Conclusions
In this study, we showed that M86 could bind to morphine-conjugated BSA or Tg but not to other related compounds conjugated to BSA or Tg. This indicated high specificity to morphine but not to codeine. SPR analysis performed using a morphine-C-Tg-coupled CM5 sensor chip showed that its Kd was 1.26 × 10−8 M. However, competitive ELISA for M86–morphine-C-Tg in the absence or presence of varying concentrations of morphine and related compounds showed binding activity with IC50 values of 257 ng/mL, 36.4, 7.3, and 7.4 nM for opium, morphine, codeine, and heroin, respectively. Thus, from a phage library constructed using mice hyperimmunized with morphine-conjugated BSA we isolated an scFv clone (M86), which reacted with free morphine, codeine, and heroin.
This study suggested limitations in the protocols of conventional animal immunization procedures and screening systems using hapten-conjugated proteins, particularly for generating small antigen-specific antibodies. However, our study results suggest that antibody engineering in combination with computer simulations is promising for developing a free morphine-specific antibody.
Thus, we established the first scFv, M86, which is a possible free morphine-specific antibody. M86 may be useful for not only detecting illegal drugs but also for developing therapeutic reagents for opiate addictions.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/2073-4468/2/1/93/s1.
Supplementary Files
Supplementary File 1Acknowledgments
We thank Masami Nakamura and Yutaka Mizuta (Central Customs Laboratory, Japan) and Satoshi Morimoto and Hiroyuki Tanaka (Department of Pharmacognosy, Graduate School of Pharmaceutical Sciences, Kyushu University, Japan) for critical discussions and for providing the narcotics.
References
- Krasowski, M.D.; Pizon, A.F.; Siam, M.G.; Giannoutsos, S.; Iyer, M.; Ekins, S. Using molecular similarity to highlight the challenges of routine immunoassay-based drug of abuse/toxicology screening in emergency medicine. BMC Emerg. Med. 2009, 9, 5. [Google Scholar]
- Baden, L.R.; Horowitz, G.; Jacoby, H.; Eliopoulos, G.M. Quinolones and false-positive urine screening for opiates by immunoassay technology. JAMA 2001, 286, 3115–3119. [Google Scholar] [CrossRef]
- Van Vunakis, H.; Wasserman, E.; Levine, L. Specificities of antibodies to morphine. J. Pharmacol. Exp. Ther. 1972, 180, 514–521. [Google Scholar]
- Glasel, J.A.; Bradbury, W.M.; Venn, R.F. Properties of murine anti-morphine antibodies. Mol. Immunol. 1983, 20, 1419–1422. [Google Scholar] [CrossRef]
- Sawada, J.; Janejai, N.; Nagamatsu, K.; Terao, T. Production and characterization of high-affinity monoclonal antibodies against morphine. Mol. Immunol. 1988, 25, 937–943. [Google Scholar] [CrossRef]
- Miller, A., 3rd; Glasel, J.A. Comparative sequence and immunochemical analyses of murine monoclonal anti-morphine antibodies. J. Mol. Biol. 1989, 209, 763–778. [Google Scholar] [CrossRef]
- Biagini, R.E.; Klincewicz, S.L.; Henningsen, G.M.; MacKenzie, B.A.; Gallagher, J.S.; Bernstein, D.I.; Bernstein, I.L. Antibodies to morphine in workers exposed to opiates at a narcotics manufacturing facility and evidence for similar antibodies in heroin abusers. Life Sci. 1990, 47, 897–908. [Google Scholar] [CrossRef]
- Lee, J.W.; Pedersen, J.E.; Moravetz, T.L.; Dzerk, A.M.; Mundt, A.D.; Shepard, K.V. Sensitive and specific radioimmunoassays for opiates using commercially available materials. I: Methods for the determinations of morphine and hydromorphone. J. Pharm. Sci. 1991, 80, 284–288. [Google Scholar] [CrossRef]
- Usagawa, T.; Itoh, Y.; Hifumi, E.; Takeyasu, A.; Nakahara, Y.; Uda, T. Characterization of morphine-specific monoclonal antibodies showing minimal cross-reactivity with codeine. J. Immunol. Methods 1993, 157, 143–148. [Google Scholar] [CrossRef]
- Sawada, J.; Yamazaki, T.; Terao, T. Molecular and biochemical analyses of combining sites of monoclonal anti-morphine antibodies. Mol. Immunol. 1993, 30, 77–86. [Google Scholar] [CrossRef]
- Yang, T.B.; Zhong, P.; Nie, J.L.; Li, J.S.; Qu, L.N.; Li, Y.H.; Kan, G.H. Preparation and identification of specific and high-affinity monoclonal antibodies against morphine. Hybridoma Hybridom. 2002, 21, 197–201. [Google Scholar] [CrossRef]
- Rahbarizadeh, F.; Rasaee, M.J.; Madani, R.; Rahbarizadeh, M.H.; Omidfar, K. Preparation and characterization of specific and high-affinity monoclonal antibodies against morphine. Hybridoma 2000, 19, 413–417. [Google Scholar] [CrossRef]
- Xi, X.; Jin, B.Q.; Zhu, Y.; Liu, X.S.; Wang, W.X.; Li, X.S.; Wu, H.Y. Preparation of monoclonal antibodies against morphine and preliminary application of its colloidal gold conjugates. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2003, 19, 395–396. [Google Scholar]
- Dillon, P.P.; Manning, B.M.; Daly, S.J.; Killard, A.J.; O’Kennedy, R. Production of a recombinant anti-morphine-3-glucuronide single-chain variable fragment (scFv) antibody for the development of a “real-time” biosensor-based immunoassay. J. Immunol. Methods 2003, 276, 151–161. [Google Scholar] [CrossRef]
- Brennan, J.; Dillon, P.; O’Kennedy, R. Production, purification and characterisation of genetically derived scFv and bifunctional antibody fragments capable of detecting illicit drug residues. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2003, 786, 327–342. [Google Scholar] [CrossRef]
- Dillon, P.P.; Daly, S.J.; Manning, B.M.; O’Kennedy, R. Immunoassay for the determination of morphine-3-glucuronide using a surface plasmon resonance-based biosensor. Biosens. Bioelectron. 2003, 18, 217–227. [Google Scholar] [CrossRef]
- Li, L.; Liu, J.; Zhu, P.; Xu, J.P.; Fu, N. Preparation of anti-morphine monoclonal antibodies with complete cross reactivity with heroin. Di Yi Jun Yi Da Xue Xue Bao 2005, 25, 833–836. [Google Scholar]
- Ma, L.X.; Zhou, Q.; Zheng, H.B.; Li, S.B. Preparation and characterization of anti-morphine vaccine antibody. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2006, 22, 368–370. [Google Scholar]
- Gandhi, S.; Caplash, N.; Sharma, P.; Raman Suri, C. Strip-based immunochromatographic assay using specific egg yolk antibodies for rapid detection of morphine in urine samples. Biosens. Bioelectron. 2009, 25, 502–505. [Google Scholar] [CrossRef]
- Pozharski, E.; Wilson, M.A.; Hewagama, A.; Shanafelt, A.B.; Petsko, G.; Ringe, D. Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine. J. Mol. Biol. 2004, 337, 691–697. [Google Scholar] [CrossRef]
- Findlay, J.W.; Butz, R.F.; Jones, E.C. Relationships between immunogen structure and antisera specificity in the narcotic alkaloid series. Clin. Chem. 1981, 27, 1524–1535. [Google Scholar]
- Moghaddam, A.; Borgen, T.; Stacy, J.; Kausmally, L.; Simonsen, B.; Marvik, O.J.; Brekke, O.H.; Braunagel, M. Identification of scFv antibody fragments that specifically recognise the heroin metabolite 6-monoacetylmorphine but not morphine. J. Immunol. Methods 2003, 280, 139–155. [Google Scholar] [CrossRef]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef]
- Kabat, E.A.; Wu, T.T. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J. Immunol. 1991, 147, 1709–1719. [Google Scholar]
- Oka, K.; Kantrowitz, J.D.; Spector, S. Isolation of morphine from toad skin. Proc. Natl. Acad. Sci. USA 1985, 82, 1852–1854. [Google Scholar] [CrossRef]
- Pelander, A.; Ojanper, I.; Hase, T.A. Preparation of N-demethylated drug metabolites for analytical purposes using 1-chloroethyl chloroformate. Forensic Sci. Int. 1997, 85, 193–198. [Google Scholar] [CrossRef]
- Burmester, J.; Plückthun, A. Construction of scFv Fragments from Hybridoma of Spleen Cells by PCR Assembly. In Antibody Engineering; Kontermann, R., Dübel, S., Eds.; Springer: Berlin, Germany, 2001; pp. 19–40. [Google Scholar]
- Hashiguchi, S.; Nakashima, T.; Nitani, A.; Yoshihara, T.; Yoshinaga, K.; Ito, Y.; Maeda, Y.; Sugimura, K. Human Fc epsilon RIalpha-specific human single-chain Fv (scFv) antibody with antagonistic activity toward IgE/Fc epsilon RIalpha-binding. J. Biochem. 2003, 133, 43–49. [Google Scholar] [CrossRef]
- Chemical Computing Group Inc. Molecular Operating Environment (MOE), 2011.10, Montreal, Canada, 2011.
- Goto, J.; Kataoka, R.; Muta, H.; Hirayama, N. ASEDock-docking based on alpha spheres and excluded volumes. J. Chem. Inf. Model. 2008, 48, 583–590. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).