2.1. Selection of sdAbs Targeting Spore Proteins
Two llamas, Whisper and Centavo, had been previously immunized with the standard veterinary vaccine for
Bacillus anthracis (Sterne 34F2 strain; Colorado Serum Company). Binding sdAbs selected from the initial library constructed following immunization with commercial vaccine primarily recognized the S-layer protein EA1, as described by Walper
et al. [
15]. Although the isolated sdAbs were specific for
B. anthracis, EA1 has been shown to be a spore contaminant and not an integral component of the bacterial spore [
17]. The two llamas were subjected to a second series of immunizations with a combination of recombinant spore-specific proteins: BclA, gerQ, SODA1, SOD15, BxpB, and p5303 [
18,
19]. Centavo received a mix of all six proteins; while Whisper was immunized with a cocktail that did not include the BclA protein in an attempt to allow a stronger immune response toward the other spore proteins that may not be as immunodominant as the BclA antigen. Plasma retained during library construction was tested from both llamas against the recombinant spore proteins as well as the EA1 protein and bacterial spores in a direct binding ELISA to assess the host llama’s immune response to the antigens,
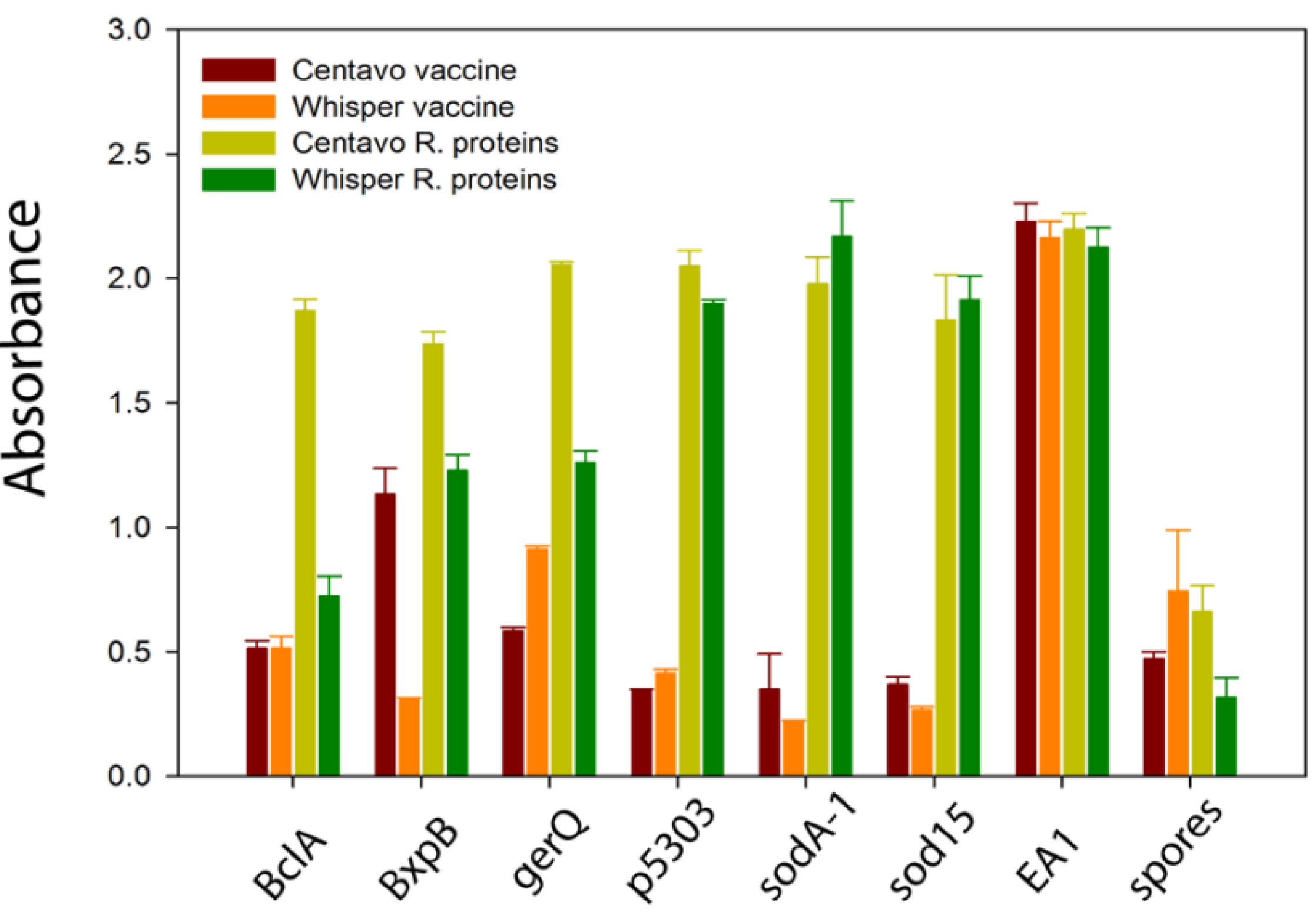
Figure 1. As anticipated, both animals demonstrated a significantly stronger immune response to the spore specific proteins following immunization with the recombinant spore proteins. Also as expected, Centavo showed a stronger response to BclA than Whisper.
Figure 1.
Binding of polyclonal antibodies from the plasma of two llamas, Centavo and Whisper, after immunization with the veterinary anthrax vaccine (red and orange) and after additional immunizations with recombinant B. anthracis spore proteins (yellow and green). Binding is shown for the six recombinant spore proteins, the EA1 S-layer protein, and B. anthracis spores.
Figure 1.
Binding of polyclonal antibodies from the plasma of two llamas, Centavo and Whisper, after immunization with the veterinary anthrax vaccine (red and orange) and after additional immunizations with recombinant B. anthracis spore proteins (yellow and green). Binding is shown for the six recombinant spore proteins, the EA1 S-layer protein, and B. anthracis spores.
The sdAb library was constructed from RNA extracted from the peripheral blood lymphocytes using a two stage PCR as outlined by Gharhroudi
et al. [
20]. Selection of target binding sdAbs was conducted using a standard biopanning protocol; two rounds of selection proved sufficient to enrich for target-binding sdAbs in all cases save BxpB, as judged by polyclonal phage ELISA,
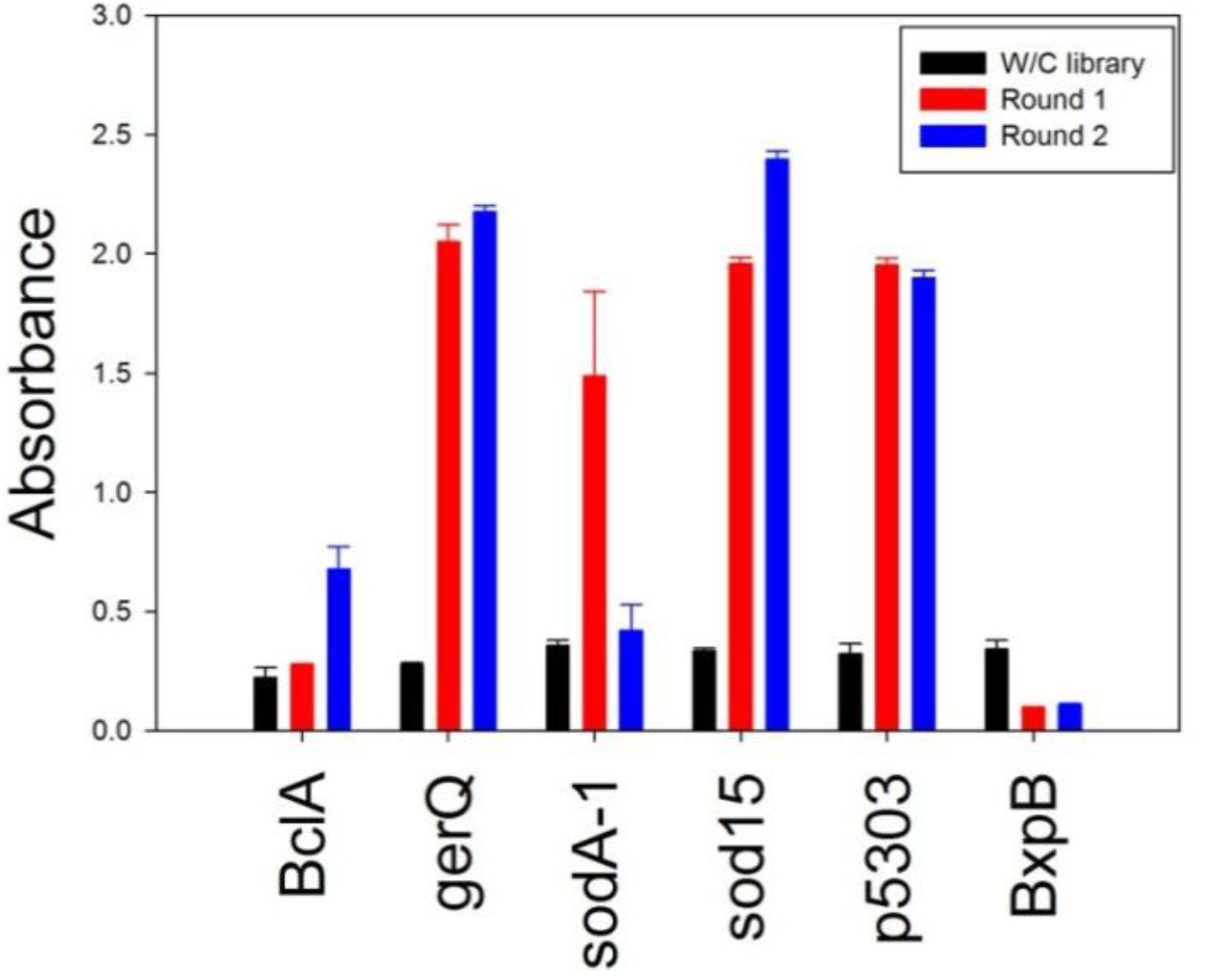
Figure 2. Though analysis of the llama plasma suggested BxpB was immunogenic, biopanning did not enrich for binders to the target protein. Biopanning was repeated for BxpB and conducted for three rounds but with similar results (data not shown). The failure to isolate BxpB binding sdAbs, despite the fact that analysis of the plasma showed binding to BxpB could be due to low diversity of the library itself. Alternatively, it is possible that the llama’s immune response may be primarily conventional antibodies rather than heavy-chain only antibodies from which the sdAb are derived. Although not done for this work, purification of the heavy-chain only antibodies is possible and can be used to assess the potential of isolating sdAbs from the library [
21].
Figure 2.
Polyclonal phage ELISA using phage prepared from the initial library and after the first and second round of biopanning. Recombinant spore proteins were passively immobilized and an HRP-conjugated anti-M13 antibody was used to assess binding.
Figure 2.
Polyclonal phage ELISA using phage prepared from the initial library and after the first and second round of biopanning. Recombinant spore proteins were passively immobilized and an HRP-conjugated anti-M13 antibody was used to assess binding.
Individual clones from the second round of biopanning for each of the six target antigens were transferred to microtiter plates and assessed for their ability to bind target using a monoclonal phage ELISA. Clones were defined as positive if the signal generated for target antigen was at least twice background. Positive clones were sequenced and divided into families based on amino acid alignments of their complementarity determining regions (CDRs). The BclA binding sdAbs exhibited the greatest diversity of sequence with eleven unique families identified. There were seven families of sdAbs selected for binding to both the SODA1 and p5303 antigens, while sdAb selected towards the SOD15 and gerQ antigens had three and four families respectively. Representative sequences from each of the binding families are shown in
Supplemental Figure 1. Though the response was minimal in the monoclonal phage ELISA (absorbance values less than two-fold greater than background), ten clones from the second round of biopanning against BxpB were also sequenced, however none were close enough in amino acid composition of the CDRs to be categorized within a defined family. These sdAbs also failed to express using a variety of vectors and expression strategies and therefore were not further characterized.
2.2. Characterization of sdAbs Targeting Spore Proteins
Representative clones from each sequence family were cloned into the pECAN45 expression vector and purified from the bacterial periplasm as detailed in Walper
et al. [
15]. Purified proteins were first tested via surface plasmon resonance (SPR) to identify those clones that recognize different epitopes on their target antigen. Despite differences in amino acid composition of the CDRs, a single epitope for each of the target antigens; or one of such proximity as to inhibit binding; was observed in all instances (data not shown). Although it is important to identify binding reagents that recognize different epitopes when performing sandwich assays for simple targets, bacterial spores are likely to have multiple copies of each of the target proteins so identifying proteins that recognize different regions is not as critical.
Following epitope evaluation, representative clones from each family were evaluated to determine binding kinetics. The majority of the clones tested showed nanomolar or better affinities for the recombinant proteins. Clones representing the two families with the highest binding affinities for each target protein were selected for further study; the binding kinetics for each is shown in
Table 1.
Table 1.
SdAb binding kinetics for recombinant proteins.
Table 1.
SdAb binding kinetics for recombinant proteins.
| sdAb clone | Ka (1/Ms) | Kd (1/s) | KD (M) |
|---|
| BclA A5 | 2.1E +05 | 1.2E -05 | 5.9E -11 |
| BclA B7 | 3.1E +05 | 2.3E -04 | 8.7E -10 |
| BclA H3 | 3.3E +05 | 5.4E -04 | 1.6E -9 |
| gerQ B2 | 1.2E+06 | 8.7E-03 | 7.6E-09 |
| gerQ G4 (to BclA) | 1.4E+05 | 2.8E+04 | 1.9E-09 |
| SODA1 F3 | 4.5E +05 | 1.4E -03 | 3.1E -09 |
| SODA1 G4 | 5.2E +05 | 1.5E -04 | 3.0E -10 |
| SOD15 A6 | 3.4E +05 | 1.7E -03 | 5.1E -09 |
| SOD15 D2 | 1.2E +05 | 1.8E -03 | 1.6E -08 |
| p5303 A3 | 1.6E +06 | 5.6E -04 | 3.6E -10 |
| p5303 B6 | 5.9E +05 | 5.5E -05 | 9.3E -11 |
| p5303 F1 | 3.2E +06 | 8.2E -03 | 2.5E -09 |
Each of the sdAbs chosen for more complete evaluation was further characterized by circular dichroism (CD) to determine its melting temperature and ability to refold following thermal denaturation. Melting temperatures (Tm) ranged from 46–72 °C with the mean Tm calculated to be 62.4 °C for the sdAbs examined,
Table 2. Though many sdAbs are able to regain their native structure following thermal denaturation, most of the sdAbs characterized via circular dichroism in this study appeared to demonstrate a propensity to misfold and aggregate during the cooling cycles. An example of an sdAb that shows good refolding on heat cycling, one that loses its ability to refold as it is heated and cooled, and one that failed to refold is shown in
Figure 3, the data from all the sdAbs evaluated for this study is shown in
Supplemental Figure 2. Several of the sdAbs did not re-fold following the initial thermal denaturation cycle as seen for all the p5303 and SOD15 binding sdAbs that were evaluated. One of the gerQ binders also showed a complete loss of structure (gerQ B2), one refolded with approximately 50% efficiency which decreases with each additional cycle (gerQ H2), and the third clone (gerQ G4) showed a 40% decrease in ellipticity following the initial cycle but appeared to refold with the same efficiency in subsequent rounds. Both of the SODA1 binders displayed an initial and consistent decrease in ellipticity with each successive round of thermal denaturation. Though BclA H3 was not able to refold after heating, both BclA A5 and BclA B7 showed significant thermal stability.
Table 2.
SdAb melting temperatures determined using circular dichroism.
Table 2.
SdAb melting temperatures determined using circular dichroism.
| SdAb clone | Temperature (°C) |
|---|
| BclA A5 | 69 |
| BclA B7 | 69 |
| BclA H3 | 58 |
| gerQ B2 | 49 |
| gerQ G4 | 46 |
| SODA1 F3 | 70 |
| SODA1 G4 | 65 |
| SOD15 A6 | 70 |
| SOD15 D2 | 65 |
| p5303 A3 | 64 |
| p5303 B6 | 72 |
| p5303 F1 | 62 |
Figure 3.
Circular dichroism of representative sdAb showing the three refolding patterns observed. BclA B7 refolds after cycles of heating and cooling; SODA1 F3 loses ability to refold during cycles of heating, and BclA H3 loses all its secondary structure after the first heating cycle.
Figure 3.
Circular dichroism of representative sdAb showing the three refolding patterns observed. BclA B7 refolds after cycles of heating and cooling; SODA1 F3 loses ability to refold during cycles of heating, and BclA H3 loses all its secondary structure after the first heating cycle.
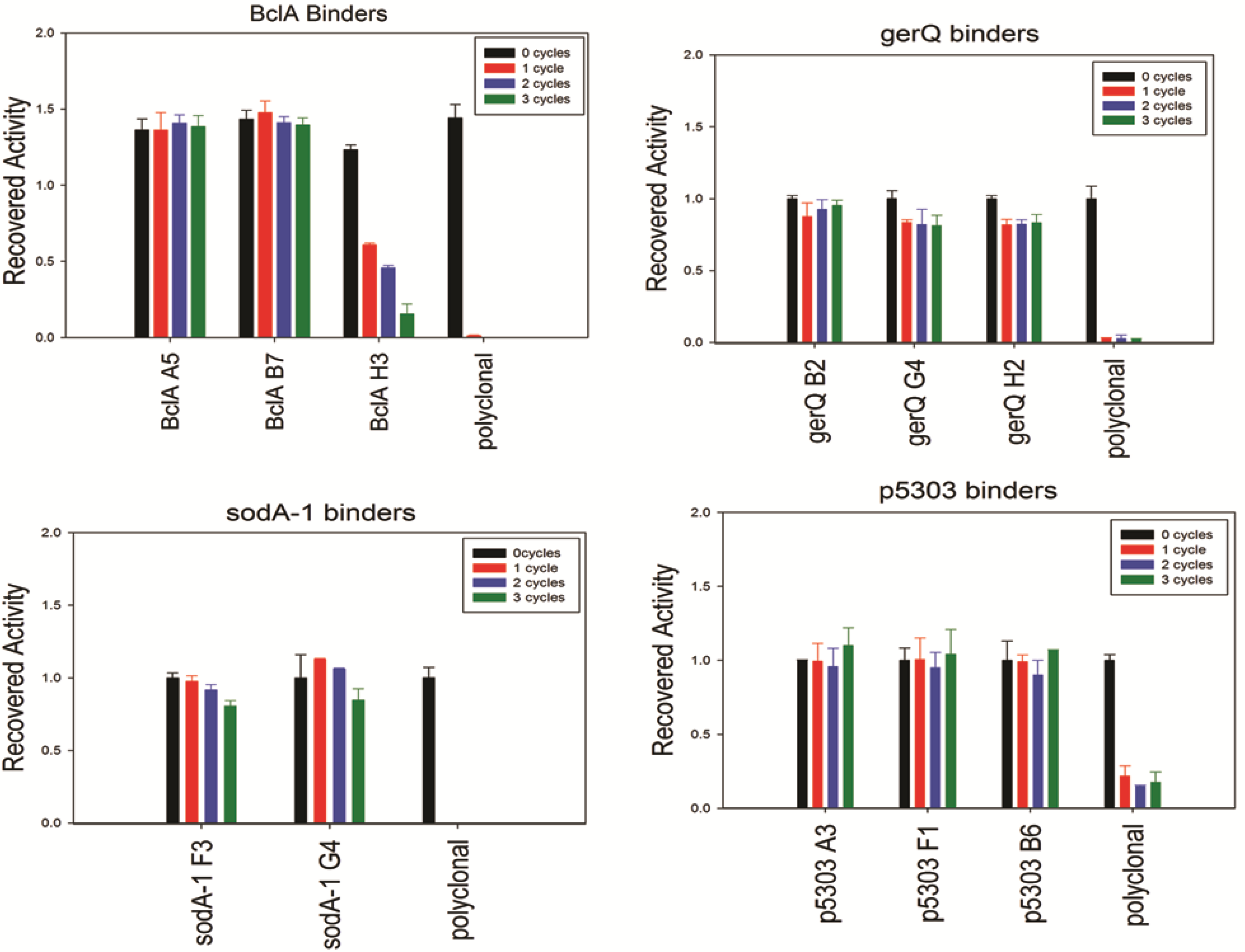
Though the CD analysis of each of the sdAbs showed significant variability in the capacity to regain secondary structure, each was tested for antigen binding capability following 0 to 3 rounds of thermal denaturation using a direct ELISA. Results are shown in
Figure 4; the sdAbs specific for SODA1, GerQ, and both BclA A5 and BclA B7 all retained an undiminished ability to bind their target antigen. Thus, while CD analysis suggests that many of these sdAb permanently lose native structure upon thermal denaturation, the direct ELISA suggests that nonetheless a sufficient percentage remains active and is able to bind the target antigen. Due to the quantity of sdAb utilized in the ELISA assay and their high affinities greater than 90% inactivation may be required before an impact is noted. In exception to the other sdAbs examined, BclA H3 showed diminished signal after each heating cycle and both of the SOD15 sdAbs were unable to reproducibly bind antigen following heating (data not shown). In all assays, the conventional polyclonal antibody that was subject to identical heating shows a near-complete loss of its ability to bind antigen following a single cycle of heating and cooling.
Figure 4.
Binding activity after iterative cycles of heating and cooling was assayed using a direct ELISA and biotinylated sdAbs. An HRP-conjugated streptavidin served as the secondary detection element.
Figure 4.
Binding activity after iterative cycles of heating and cooling was assayed using a direct ELISA and biotinylated sdAbs. An HRP-conjugated streptavidin served as the secondary detection element.
2.3. Detection of Spores (Broken and Intact)
The ability to discriminate between the spores of
Bacillus anthracis and other
Bacillus species is of significant importance for the development of any assay or sensor. As an initial test of target specificity we evaluated the ability of the sdAb to recognize spores from
B. anthracis Sterne and a limited number of closely related Bacillus strains. Spores were harvested from
B. anthracis Sterne strain 34F2,
B.cereus 13061,
B. mycoides,
B. subtilis, and
B. thuringiensis Kurstaki 4D9 as described in Walper
et al. [
15]. Purified spores were mechanically sheared using 1.0 mm glass beads, a BioSpec Products Mini-Bead Beater, and a protocol modified from that described by Vandeventer
et al. [
22]. Protein concentrations obtained for several of the spore samples, particularly
B. mycoides and
B. subtilis, were significantly lower (10-fold) than those obtained from the other spore samples. Rather than immobilize spore material based on original spore optical density, equivalent protein concentrations were immobilized to microtiter plates. This was done to ensure that a negative result was not a result of low protein concentration rather an indicator of specificity. The sdAbs targeting SOD15 and p5303 failed to generate a positive signal on any of the spore material using a direct ELISA method. In contrast, the sdAbs selected for binding spore proteins BclA, gerQ, and SODA1 all showed the ability to bind immobilized spore material and to discriminate between the spores of
B. anthracis and the spore material of non-target
Bacillus species,
Figure 5 and
Supplemental Figure 3. While the signal generated by most of the spore-specific sdAbs was minimal, a positive signal was evident above background. This could be observed within five minutes of substrate addition. In contrast to other spore-specific protein binders, the BclA binder A5 generated a significant signal for target spore material, comparable to the signal generated by the EA1 binding sdAb BA G10.
Figure 5.
Specificity assay. Direct binding ELISA of the isolated sdAb to broken spores from B. anthracis Sterne and other Bacillus species. Binding of biotinylated sdAbs was detected with an HRP-conjugated streptavidin.
Figure 5.
Specificity assay. Direct binding ELISA of the isolated sdAb to broken spores from B. anthracis Sterne and other Bacillus species. Binding of biotinylated sdAbs was detected with an HRP-conjugated streptavidin.
It has been shown previously that antibodies can readily recognize broken spore material; difficulties, however, can be encountered when attempting to detect intact bacterial spores [
15,
23]. As an intact structure, the proteins of the exosporium and coat likely occlude antibody epitopes that are readily exposed once the spore is broken. Intact
B. anthracis Sterne strain spores were washed repeatedly to remove debris then immobilized to microtiter plates as described above. With the exception of the BclA A5 and BA G10 sdAbs, intact spore detection was unsuccessful using this assay format,
Figure 6. The absorbance measured for BclA A5 was threefold lower at the highest concentration compared to BA G10 and twofold lower than the polyclonal antibody.
Figure 6.
Limit of detection for intact B. anthracis Sterne strain spores assessed using a direct binding ELISA assay.
Figure 6.
Limit of detection for intact B. anthracis Sterne strain spores assessed using a direct binding ELISA assay.
A series of bead-based, sandwich assays were conducted using the MAGPIX system to assess limits of detection for both broken and intact spores. Prior to developing the sandwich assays, an initial assay was conducted in which recombinant target protein was immobilized to magnetic beads to better evaluate specificity of individual sdAbs,
Supplemental Figure 4. In this assay, many of the sdAbs showed significant levels of cross reactivity in particular with the recombinant BclA antigen which has consistently exhibited a characteristic “stickiness”. This is typified by BA G10, which showed low affinity binding to BclA, but high affinity binding to its target EA1 (
Supplemental Figure 4, panel B). Though unexpected, these results were confirmed using a direct binding ELISA (data not shown). In the direct binding MAGPIX assays, the critical factor is the antibody titer more than absolute signal level; the amount of active antigen on each type of bead set may vary. However it is encouraging when both high titer and high signal are observed. Examining this data, it appears that at least in one case the sdAb’s target was misidentified, as the gerQ binder G4 binds BclA with higher affinity than it does gerQ, while the gerQ binder B2 had a similar affinity for both gerQ and BclA. These results were confirmed using an SPR chip that had all spore coat protein immobilized. The sdAb showing the best affinity and specificity were the BclA binders A5 and B7, and the p5303 binders A3 and F1.
Seven of the newly isolated sdAbs, along with BA G10 and polyclonal goat anti-
B. anthracis antibody, were directly immobilized to magnetic bead sets to serve as captures in sandwich assays. The biotinylated sdAbs used in the ELISA and MAGPIX specificity assays served as detection antibodies to assess limits of detection for both broken and intact spore samples. Purified
B. anthracis spores were diluted in PBS then either broken using glass beads or used intact in sandwich MAGPIX assays.
Figure 7 shows four representative assays for intact BA spores using the sdAbs BclA A5, SODA1 G4, p5303 A3, and polyclonal goat anti-
anthracis as the biotinylated detection antibodies in conjunction with all nine capture reagents; the data for all the sandwich assays are shown in
Supplemental Figure 5.
Figure 7.
Sandwich immunoassays using four tracers in combination with nine capture reagents for the detection of broken (top) and intact (bottom) B. anthracis spores. Seven of the spore specific sdAbs, BA G10 (a previously isolated anti-B. anthracis sdAb), and a polyclonal goat anti-anthrax antibody were used as captures. The X-axis defines the approximate spore concentration (cfu/mL). The highest concentration (1 × 106 spores/mL), an intermediate concentration, and the lowest concentration (no spores) is shown for each data set. Other data points represent a 1:5 serial dilution. Labels were omitted to improve clarity of the axis.
Figure 7.
Sandwich immunoassays using four tracers in combination with nine capture reagents for the detection of broken (top) and intact (bottom) B. anthracis spores. Seven of the spore specific sdAbs, BA G10 (a previously isolated anti-B. anthracis sdAb), and a polyclonal goat anti-anthrax antibody were used as captures. The X-axis defines the approximate spore concentration (cfu/mL). The highest concentration (1 × 106 spores/mL), an intermediate concentration, and the lowest concentration (no spores) is shown for each data set. Other data points represent a 1:5 serial dilution. Labels were omitted to improve clarity of the axis.
Surprisingly, the anti-p5303 sdAbs, which had not shown binding to spores in direct ELISA assays provided detection of intact spores down to at least 1.23 × 10
4 spores/mL in the MAGPIX sandwich assay. As anticipated, the most successful tracer varied with each capture molecule for the detection of both broken and intact spore samples. Regardless of the combination of capture and tracer, intact spores were reliably detected with the best capture: tracer pairs to approximately 1.23 × 10
4 spores/mL and broken spores to about 4.12 × 10
3 spores/mL as shown in
Supplemental Figure 4. The lowest limit of detection was observed when the affinity purified polyclonal goat anti-
B. anthracis antibody was used as the capture and polyclonal goat anti-
B. anthracis antibody as the tracer antibody. Several of the sdAb tracers gave the best detection when paired with the polyclonal capture. However, effective sdAb pairs for the detection of both broken and intact spores were also observed. For example, the sdAb BclA B7 capture showed good detection of intact spores when it was paired with the SODA1 binding sdAb F3 or the gerQ binding sdAb B2 as the tracers. Similarly, the sdAbs binding the putative spore protein p5303 (A3, B6, and F1) provided good detection when used in combination as capture and tracer pairs. An order of magnitude improvement in limit of detection was observed when broken spore material was used in these assays,
Supplemental Figure 5.
As detailed above, the sdAbs were not able to achieve the limits of detection shown by the conventional antibodies when used as either captures or tracers in MAGPIX assays. This is likely not the result of poor binding, rather the small size of the sdAb can result in a degree of inactivation upon immobilization when utilized as the capture reagent and a reduced level of attached biotin when utilized as the recognition reagent, both these factors can contribute to reduced signal amplification. While not evaluated for these particular clones, the sdAb platform is amenable to the addition of fusion proteins that allow for not only an increase in available biotinylation sites but also the development of assays in which the sdAb directly serves as the reporter [
24]. Future development of such assays may better showcase the functionality of sdAbs in spore detection assays.