Bio-Based Decontamination and Detoxification of Total Petroleum Hydrocarbon-Contaminated Dredged Sediments: Perspectives to Produce Constructed Technosols in the Frame of the Circular Economy
Abstract
:1. Introduction
2. Materials and Methods
2.1. TPH-Contaminated Co-Composting Sediments,, Fungal Strain, and Chemicals
2.2. Mesocosms-Scale Experimentation
2.3. Quantification of TPH, Humic and Fulvic Acids, and Ergosterol
2.4. Metabarcoding Analysis
2.5. Data Analysis
2.6. Cell Proliferation Assay
2.7. Micronucleus Frequency Assay
3. Results
3.1. Quantification of the Total Petroleum Hydrocarbons, Humic and Fulvic Acids, and Ergosterol Content in Mesocosms
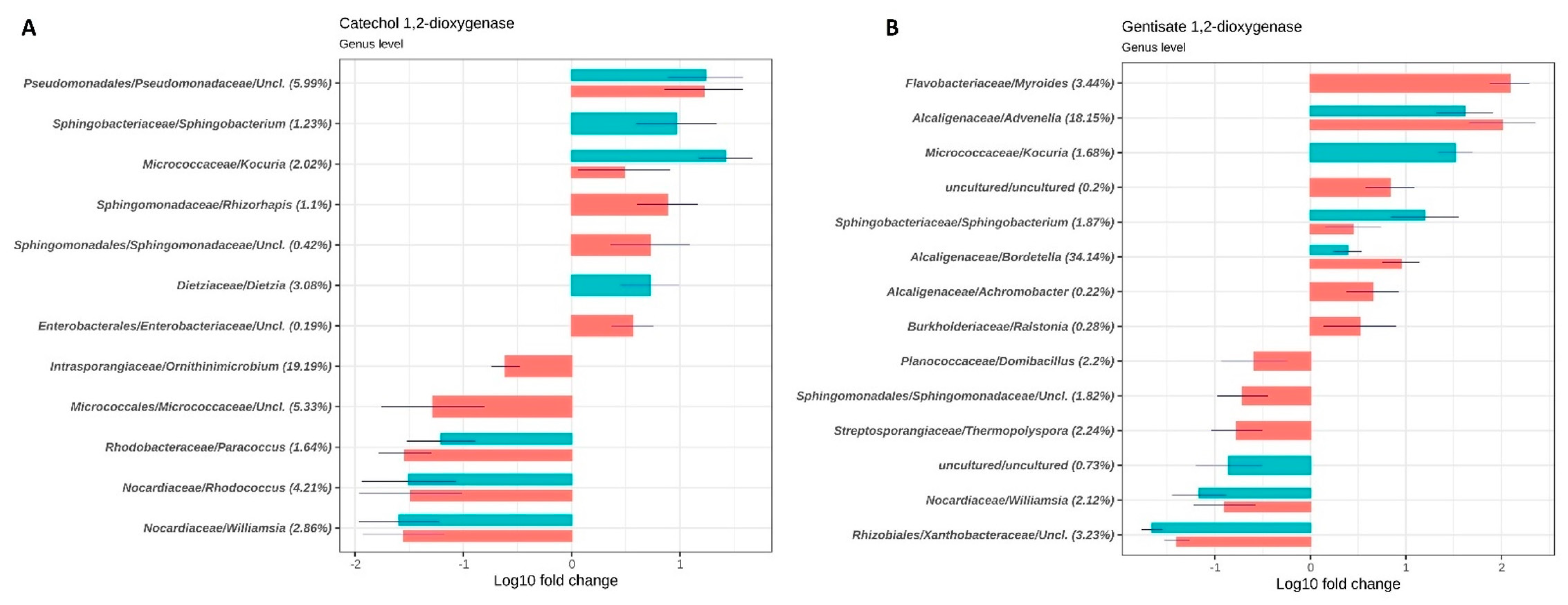
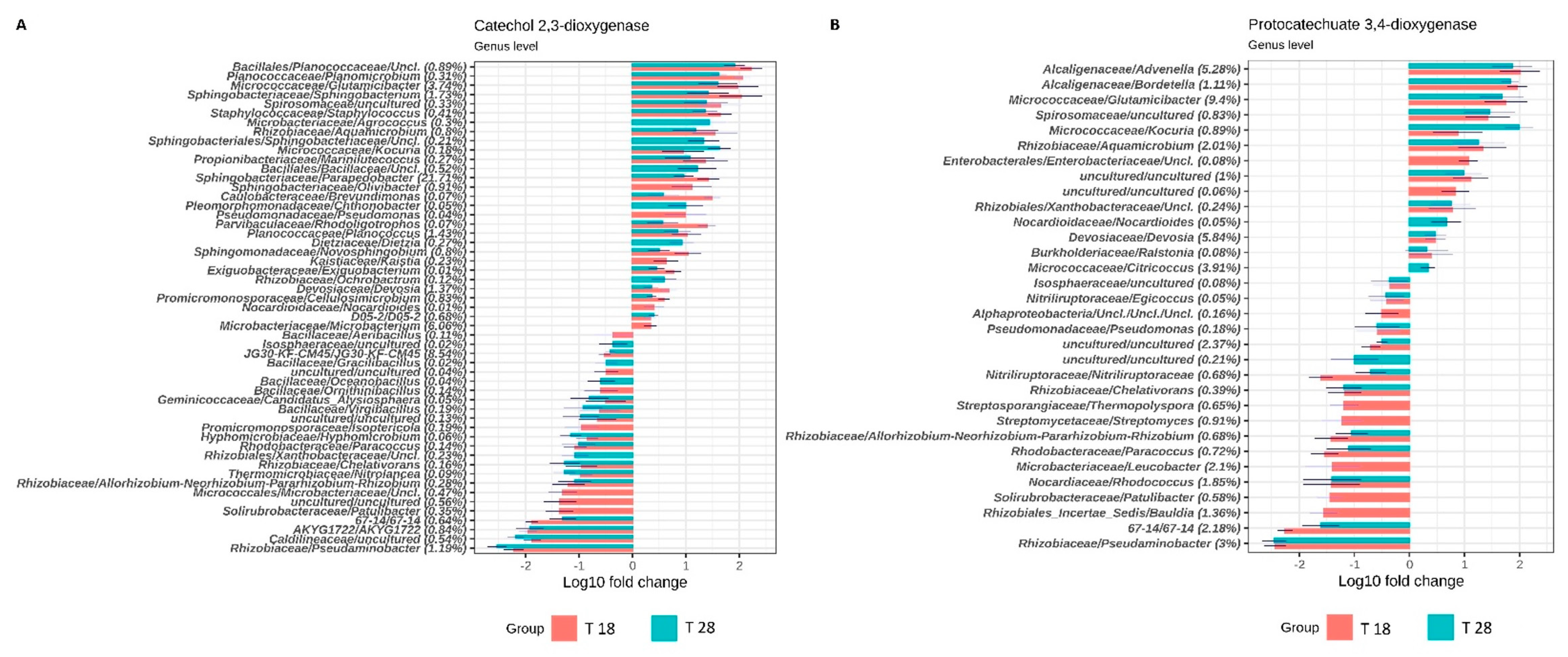
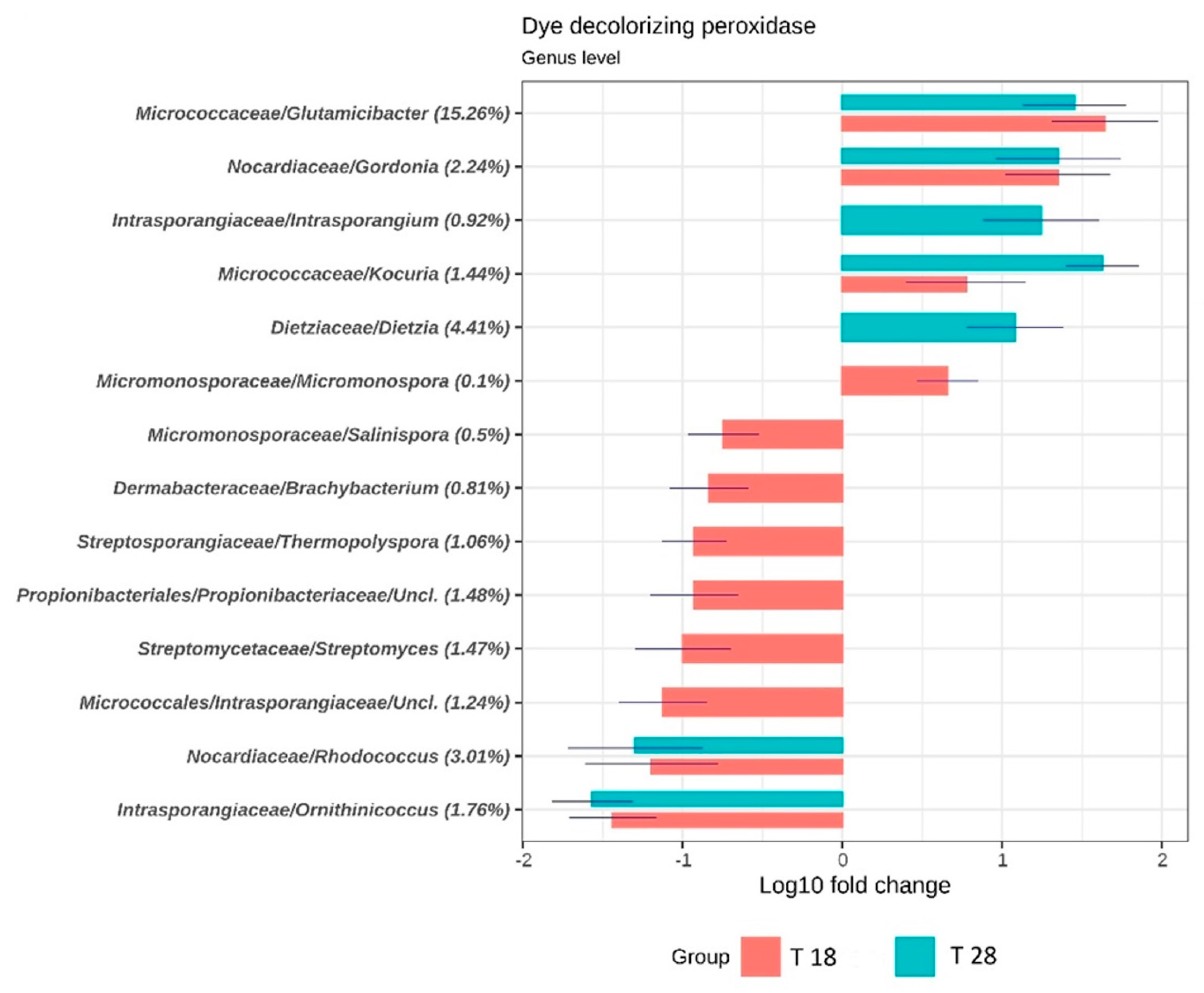
3.2. Taxonomic and Functional Analysis
3.3. Plant Bioassay
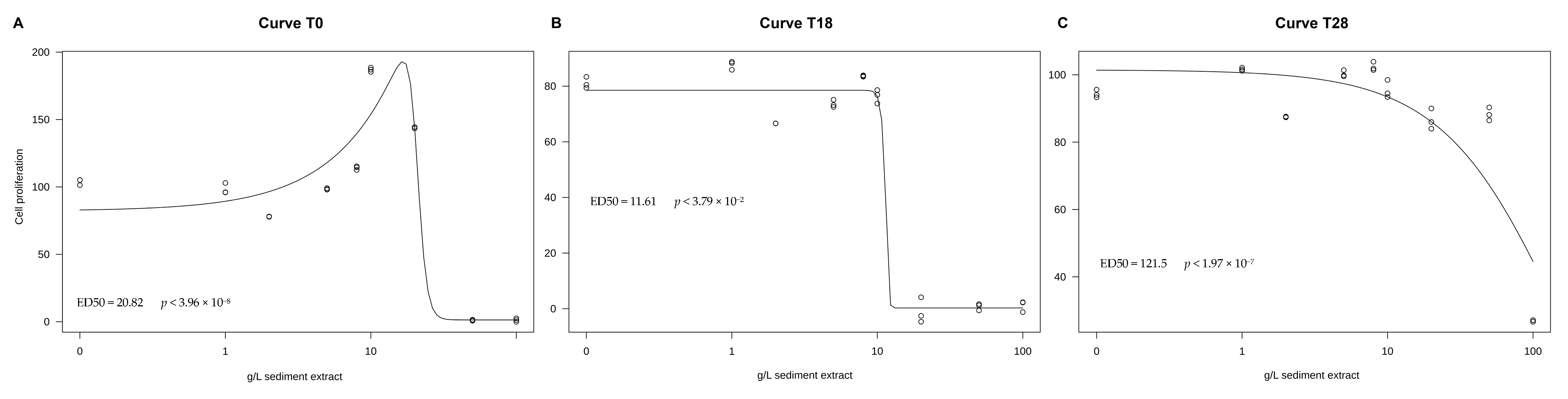
3.4. CaCo-2 Cell Proliferation Assay
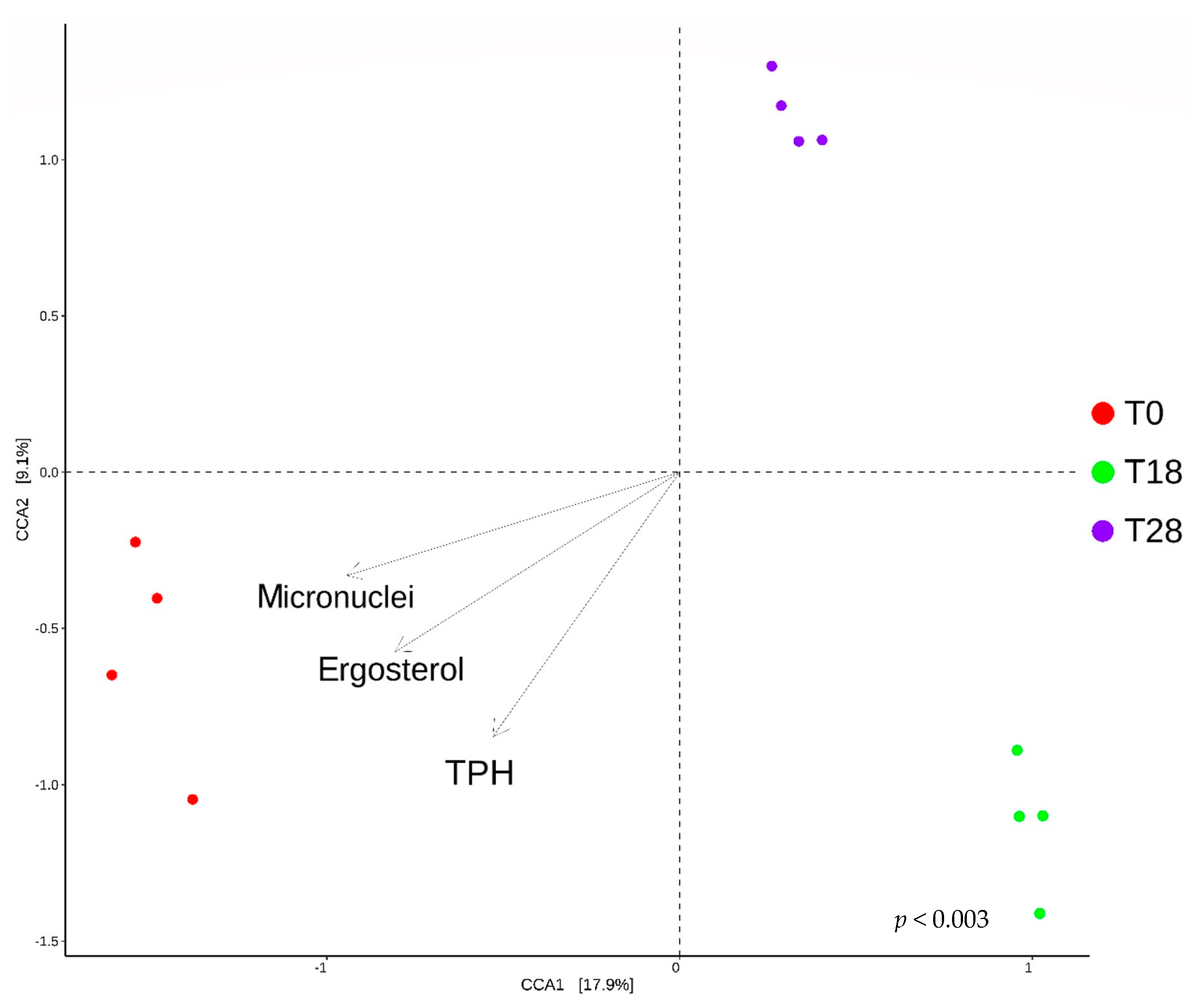
3.5. Canonical Correspondence Analysis of the Process
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- IUSS Working Group WRB. World Reference Base for Soil Resources 2014, Update 2015; International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; World Soil Resources Reports No. 106; FAO: Rome, Italy, 2015. [Google Scholar]
- Margulis, M.E.; McKeon, N.; Borras, S., Jr. Land Grabbing and Global Governance: Critical Perspectives. Globalizations 2013, 10, 1–23. [Google Scholar] [CrossRef]
- Moraga, G.; Huysveld, S.; Mathieux, F.; Blengini, G.A.; Alaerts, L.; Van Acker, K.; de Meester, S.; Dewulf, J. Circular economy indicators: What do they measure? Resour. Conserv. Recycl. 2019, 146, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Cascone, S. Green Roof Design: State of the Art on Technology and Materials. Sustainability 2019, 11, 3020. [Google Scholar] [CrossRef]
- Dell’Anno, A.; Beolchini, F.; Corinaldesi, C.; Amato, A.; Becci, A.; Rastelli, E.; Hekeu, M.; Regoli, F.; Astarita, E.; Greco, S.; et al. Assessing the efficiency and eco-sustainability of bioremediation strategies for the reclamation of highly contaminated marine sediments. Mar. Environ. Res. 2020, 162, 105101. [Google Scholar] [CrossRef] [PubMed]
- Becarelli, S.; Chicca, I.; Siracusa, G.; La China, S.; Gentini, A.; Lorenzi, R.; Munz, G.; Petroni, G.; Levin, D.B.; Di Gregorio, S. Hydrocarbonoclastic Ascomycetes to enhance co-composting of total petroleum hydrocarbon (TPH) contaminated dredged sediments and lignocellulosic matrices. N. Biotechnol. 2019, 50, 27–36. [Google Scholar] [CrossRef]
- Becarelli, S.; Chicca, I.; La China, S.; Siracusa, G.; Bardi, A.; Gullo, M.; Petroni, G.; Levin, D.B.; Di Gregorio, S. A New Ciboria sp. for Soil Mycoremediation and the Bacterial Contribution to the Depletion of Total Petroleum Hydrocarbons. Front. Microbiol. 2021, 12, 647373. [Google Scholar] [CrossRef]
- Di Gregorio, S.; Siracusa, G.; Becarelli, S.; Mariotti, L.; Gentini, A.; Lorenzi, R. Isolation and characterization of a hydrocarbonoclastic bacterial enrichment from total petroleum hydrocarbon contaminated sediments: Potential candidates for bioaugmentation in bio-based processes. Environ. Sci. Pollut. Res. Int. 2016, 23, 10587–10594. [Google Scholar] [CrossRef]
- Riser-Roberts, E. Remediation of Petroleum Contaminated Soil: Biological, Physical, and Chemical Processes; Lewis Publishers: Boca Raton, FL, USA, 1998. [Google Scholar]
- Ruffini Castiglione, M.; Giorgetti, L.; Becarelli, S.; Siracusa, G.; Lorenzi, R.; Di Gregorio, S. Polycyclic aromatic hydrocarbon-contaminated soils: Bioaugmentation of autochthonous bacteria and toxicological assessment of the bioremediation process by means of Vicia faba L. Environ. Sci. Pollut. Res. Int. 2016, 23, 7930–7941. [Google Scholar] [CrossRef]
- ISO 29200:2013; Soilquality—Assessment of Genotoxicity Effects on Higher Plants—Vicia faba Micreonucleus Test. ISO: Geneva, Switzerland, 2013.
- Balimane, P.V.; Chong, S.; Morrison, R.A. Current methodologies used for evaluation of intestinal permeability and absorption. J. Pharmacol. Toxicol. Methods 2000, 44, 301–312. [Google Scholar] [CrossRef]
- Becarelli, S.; Siracusa, G.; Chicca, I.; Bernabei, G.; Di Gregorio, S. Ascomycetes versus Spent Mushroom Substrate in Mycoremediation of Dredged Sediments Contaminated by Total Petroleum Hydrocarbons: The Involvement of the Bacterial Metabolism. Water 2021, 13, 3040. [Google Scholar] [CrossRef]
- Buiarelli, F.; Canepari, S.; Di Filippo, P.; Perrino, C.; Pomata, D.; Riccardi, C.; Speziale, R. Extraction and analysis of fungal spore biomarkers in atmospheric bioaerosol by HPLC-MS-MS and GC-MS. Talanta 2013, 105, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Olsson, P.A.; Larsson, L.; Bago, B.; Wallander, H.; Van Aarle, I.M. Ergosterol and fatty acids for biomass estimation of mycorrhizal fungi. N. Phytol. 2003, 159, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physi. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Shrestha, R.; Jia, K.; Gao, P.F.; Geisbrecht, B.V.; Bossmann, S.H.; Shi, J.; Li, P. Characterization of Dye-decolorizing Peroxidase (DyP) from Thermomonospora curvata Reveals Unique Catalytic Properties of A-type DyPs. J. Biol. Chem. 2015, 290, 23447–23463. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, T. Bacterial dye-decolorizing peroxidases: Biochemical properties and biotechnological opportunities. Phys. Sci. Rev. 2016, 1, 20160051. [Google Scholar]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Peddada, S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef]
- Pérez-Pantoja, D.; Donoso, R.; Agulló, L.; Córdova, M.; Seeger, M.; Pieper, D.H.; González, B. Genomic analysis of the potential for aromatic compounds biodegradation in Burkholderiales. Environ. Microbiol. 2012, 14, 1091–1117. [Google Scholar] [CrossRef]
- Hopper, D.J.; Taylor, D.G. Pathways for the degradation of m-cresol and p-cresol by Pseudomonas putida. J. Bacteriol. 1975, 122, 1–6. [Google Scholar] [CrossRef]
- Li, W.; Shi, J.; Wang, X.; Han, Y.; Tong, W.; Ma, L.; Liu, B.; Cai, B. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 2004, 336, 231–240. [Google Scholar] [CrossRef]
- Shen, X.; Liu, S. Key enzymes of the protocatechuate branch of the beta-ketoadipate pathway for aromatic degradation in Corynebacterium glutamicum. Sci. China C Life Sci. 2005, 48, 241–249. [Google Scholar] [PubMed]
- Ferraro, D.J.; Gakhar, L.; Ramaswamy, S. Rieske business: Structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 2005, 338, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Scarlett, A.; Galloway, T.; Rowland, S. Chronic toxicity of unresolved complex mixtures (UCM) of hydrocarbons in marine sediments. J. Soil. Sediment. 2007, 7, 200–206. [Google Scholar] [CrossRef]
- Thomas, K.V.; Donkin, P.; Rowland, S.J. Toxicity enhancement of an aliphatic petrogenic unresolved complex mixture (UCM) by chemical oxidation. Water Res. 1995, 29, 379–382. [Google Scholar] [CrossRef]
- Booth, A.M.; Sutton, P.A.; Lewis, C.A.; Scarlett, A.; Chau, W.; Widdows, J.; Rowland, S.J. Un-resolved complex mixtures of aromatic hydrocarbons: Thousands of overlooked persistent, bioaccumulative, and toxic contaminants in mussels. Environ. Sci. Technol. 2007, 41, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Booth, A.M.; Scarlett, A.G.; Lewis, C.A.; Belt, S.T.; Rowland, S.J. Unresolved complex mixtures (UCMs) of aromatic hydrocarbons: Branched alkyl indanes and branched alkyl tetralins are present in UCMs and accumulated by and toxic to, the mussel Mytilus edulis. Environ. Sci. Technol. 2008, 42, 8122–8126. [Google Scholar] [CrossRef]
- Samanipoura, S.; Dimitriou-Christidisa, P.; Grosa, J.; Grangea, A.; Samuel, A.J. Analyte quantification with comprehensive two-dimensional gas chromatography: Assessment of methods for baseline correction, peak delineation, and matrix effect elimination for real samples. J. Chromatogr. A 2015, 1375, 123–139. [Google Scholar] [CrossRef]
- Li, F.; Guo, S.; Hartog, N.; Yuan, Y.; Yang, X. Isolation and characterization of heavy polycyclic aromatic hydrocarbon-degrading bacteria adapted to electrokinetic conditions. Biodegradation 2016, 27, 1–13. [Google Scholar] [CrossRef]
- Hidalgo, K.J.; Sierra-Garcia, I.N.; Dellagnezze, B.M.; de Oliveira, V.M. Metagenomic Insights into the Mechanisms for Biodegradation of Polycyclic Aromatic Hydrocarbons in the Oil Supply Chain. Front. Microbiol. 2020, 11, 561506. [Google Scholar] [CrossRef]
- Eriksson, M.; Sodersten, E.; Yu, Z.; Dalhammar, G.; Mohn, W.W. Degradation of polycyclic aromatic hydrocarbons at low temperature under aerobic and nitrate-reducing conditions in enrichment cultures from northern soils. Appl. Environ. Microbiol. 2003, 69, 275–284. [Google Scholar] [CrossRef]
- Wang, X.; Jin, D.; Zhou, L.; Wu, L.; An, W.; Zhao, L. Draft Genome Sequence of Advenella kashmirensis Strain W13003, a Polycyclic Aromatic Hydrocarbon-Degrading Bacterium. Genome Announc. 2014, 2, e00003-14. [Google Scholar] [CrossRef] [PubMed]
- Maneerat, S.; Bamba, T.; Harada, K.; Kobayashi, A.; Yamada, H.; Kawai, F. A novel crude oil emulsifier excreted in the culture supernatant of a marine bacterium, Myroides sp. strain SM1. Appl. Microbiol. Biotechnol. 2006, 70, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zheng, G.; Zhou, W.; Wang, J.; Zhou, L. Bioleaching conditioning increased the bioavailability of polycyclic aromatic hydrocarbons to promote their removal during co-composting of industrial and municipal sewage sludges. Sci. Total Environ. 2019, 665, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Guo, X.; Wu, J.; Chen, K. Effect of compost amendment and bioaugmentation on PAH degradation and microbial community shifting in petroleum-contaminated soil. Chemosphere 2020, 256, 126998. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Niu, X.; Tian, Y.; Xiao, Y. Assessment of PAH degradation potential of native species from a coking plant through identifying of the beneficial bacterial community within the rhizosphere soil. Chemosphere 2021, 264, 128513. [Google Scholar] [CrossRef] [PubMed]
- Andreoni, V.; Cavalca, L.; Rao, M.A.; Nocerino, G.; Bernasconi, S.; Dell’Amico, E.; Colombo, M.; Gianfreda, L. Bacterial communities and enzyme activities of PAHs polluted soils. Chemosphere 2004, 57, 401–412. [Google Scholar] [CrossRef]
- Zavarzina, A.G.; Lisov, A.A.; Zavarzin, A.A.; Leontievsky, A.A. Fungal oxidoreductases and humification in forest soils. In Soil Enzymology; Shukla, G., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 207–228. [Google Scholar]
- Tran, H.T.; Lin, C.; Bui, X.T.; Ngo, H.H.; Cheruiyot, N.K.; Hoang, H.G.; Vu, C.T. Aerobic composting remediation of petroleum hydrocarbon-contaminated soil. Current and future perspectives. Sci. Total Environ. 2021, 753, 142250. [Google Scholar] [CrossRef]
- Wong, D.W.S. Structure and action mechanism of ligninolytic enzymes. Appl. Biochem. Biotechnol. 2008, 157, 174–209. [Google Scholar] [CrossRef]
- Huang, M.; Zhu, Y.; Li, Z.; Huang, B.; Luo, N.; Liu, C.; Zeng, G. Compost as a soil amendment to remediate heavy metal-contaminated agricultural soil: Mechanisms, efficacy, problems, and strategies. Water Air Soil Pollut. 2016, 227, 359. [Google Scholar] [CrossRef]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Vokou, D.; Karamanoli, K.; Mellidou, I. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 2020, 10, 14857. [Google Scholar] [CrossRef]
- Chen, M.; Li, N.; Zhang, X.F.; Zhou, X.K.; Shi, R.; Su, Y.X.; Liu, J.J.; Cao, Y.; Mo, M.H.; Ma, L. Sphingobacterium faecale sp. nov., a 1-aminocyclopropane-1-carboxylate deaminase producing bacterium isolated from camel faeces. Int. J. Syst. Evol. Microbiol. 2022, 72, 5215. [Google Scholar] [CrossRef] [PubMed]
- Gholami, M.; Etemadifar, Z.; Bouzari, M. Isolation a new strain of Kocuria rosea capable of tolerating extreme conditions. J. Environ. Radioact. 2015, 144, 113–119. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Niu, Z.; Liu, T.; Su, Y. Succession of soil bacterial community along a 46-year choronsequence artificial revegetation in an arid oasis-desert ecotone. Sci. Total Environ. 2022, 814, 152496. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Berbel, N.; Soria, R.; Ortega, R.; Bastida, F.; Miralles, I. Quarry restoration treatments from recycled waste modify the physicochemical soil properties, composition and activity of bacterial communities and priming effect in semi-arid areas. Sci. Total Environ. 2021, 774, 145693. [Google Scholar] [CrossRef]
- Zhang, S.; Fang, Y.; Luo, Y.; Li, Y.; Ge, T.; Wang, Y.; Wang, H.; Yu, B.; Song, X.; Chen, J.; et al. Linking soil carbon availability, microbial community composition and enzyme activities to organic carbon mineralization of a bamboo forest soil amended with pyrogenic and fresh organic matter. Sci. Total Environ. 2021, 801, 149717. [Google Scholar] [CrossRef]




| Components | Dredged Sediments |
|---|---|
| Total Petroleum Hydrocarbons | 2738 ± 79 mg/kg |
| Total Phosphate | 15.0 ± 0.2 mg/kg |
| Total Nitrogen | 0.31 ± 0.02 mg/kg |
| Chloride | 2400 ± 2 mg/kg |
| Pb | 21.0 ± 0.1 mg/kg |
| Cd | 15.0 ± 0.2 mg/kg |
| Zn | 7.0 ± 0.1 mg/kg |
| Cr | 9.0 ± 0.2 mg/kg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becarelli, S.; Bernabei, G.; Siracusa, G.; Baderna, D.; Ruffini Castiglione, M.; De Simone, G.; Di Gregorio, S. Bio-Based Decontamination and Detoxification of Total Petroleum Hydrocarbon-Contaminated Dredged Sediments: Perspectives to Produce Constructed Technosols in the Frame of the Circular Economy. Water 2023, 15, 4106. https://doi.org/10.3390/w15234106
Becarelli S, Bernabei G, Siracusa G, Baderna D, Ruffini Castiglione M, De Simone G, Di Gregorio S. Bio-Based Decontamination and Detoxification of Total Petroleum Hydrocarbon-Contaminated Dredged Sediments: Perspectives to Produce Constructed Technosols in the Frame of the Circular Economy. Water. 2023; 15(23):4106. https://doi.org/10.3390/w15234106
Chicago/Turabian StyleBecarelli, Simone, Giacomo Bernabei, Giovanna Siracusa, Diego Baderna, Monica Ruffini Castiglione, Giampiero De Simone, and Simona Di Gregorio. 2023. "Bio-Based Decontamination and Detoxification of Total Petroleum Hydrocarbon-Contaminated Dredged Sediments: Perspectives to Produce Constructed Technosols in the Frame of the Circular Economy" Water 15, no. 23: 4106. https://doi.org/10.3390/w15234106









