Abstract
Enterovirus 70 (EV70) is an emerging viral pathogen that remains viable in final treated effluent. Solar irradiation is, therefore, explored as a low-cost natural disinfection strategy to mitigate potential concerns. EV70 was exposed to simulated sunlight for 24 h at a fluence rate of 28.67 J/cm2/h in three different water matrices, namely, phosphate-buffered saline (PBS), treated wastewater effluent, and chlorinated effluent. In the presence of sunlight, EV70 decreased in infectivity by 1.7 log, 1.0 log, and 1.3 log in PBS, effluent, and chlorinated effluent, respectively. Irradiated EV70 was further introduced to host cell lines and was unable to infect the cell lines. In contrast, EV70 in dark microcosms replicated to titers 13.5, 3.3, and 4.2 times the initial inoculum. The reduction in EV70 infectivity was accompanied by a reduction in viral binding capacity to Vero cells. In addition, genome sequencing analysis revealed five nonsynonymous nucleotide substitutions in irradiated viruses after 10 days of infection in Vero cells, resulting in amino acid substitutions: Lys14Glu in the VP4 protein, Ala201Val in VP2, Gly71Ser in VP3, Glu50Gln in VP1, and Ile47Leu in 3Cpro. Overall, solar irradiation resulted in EV70 inactivation and an inhibition of viral activity in all parameters studied.
1. Introduction
Climate change, urbanization, and increasing global population have placed considerable pressure on freshwater supplies [1,2,3]. Wastewater can be used as an alternative water resource for agriculture irrigation and aquifer recharge but would first require appropriate treatment in wastewater treatment plants (WWTPs). WWTPs act as engineered barriers to treat municipal wastewater to a quality that is sufficiently safe for reuse. In most WWTPs, the final treatment step typically includes the use of chlorine as a disinfectant to reduce the biological activity of remnant pathogens present in the treated effluent [4]. However, each pathogen reacts differently to different disinfectants, and a single disinfection strategy is rarely effective against all pathogens [5]. For example, a WWTP utilizing chlorination as a disinfection strategy was able to inactivate human adenoviruses but not enteroviruses fully from wastewater [6]. Additional disinfection strategies, particularly those that are low-cost and easily accessible, may have to be deployed to further inactivate remnant viral contaminants.
Solar irradiation is a freely accessible, low-cost biocidal strategy that is abundant in many tropical countries and can be used to circumvent this need. The biocidal effect of sunlight works through the effects of ultraviolet A (UV-A) and ultraviolet B (UV-B). UV-A, of wavelengths 320–400 nm, is absorbed by molecular chromophores which, in turn, generate reactive oxidative species (ROS). ROS induce damage to cellular membranes, proteins and nucleic acids, rendering viruses and other pathogens inactive. UV-B, of wavelengths 280–320 nm, functions directly through absorption by nucleic acids and proteins. UV-B can also affect pyrimidines directly, inducing mutagenic and genotoxic effects in the genomes of microbes [7,8].
Several studies have documented the effects of irradiation on viruses. However, the dosage required for viral inactivation varies widely with viral species, particle size, genome type, length, and polarity [9,10,11,12]. For instance, numerous studies have investigated the effects of solar irradiation on members of the Picornaviridae family, which contain a single positively stranded RNA genome [13]. Heaselgrave et al., reported a 4-log inactivation of polioviruses with solar irradiation ranging from 198 to 1224 J/cm2 [14,15]. In contrast, Coxsackie viruses required 117–198 J/cm2 of solar irradiation for a 4-log inactivation, while ECHO viruses required 50–60 J/cm2 for a 2-log reduction [15,16].
The variation in solar intensity required to inactivate different RNA viruses within the same family shows that susceptibility of viruses to solar irradiation differs at the species level. A species within the Picornaviridae family that has not been studied in this aspect is enterovirus 70 (EV70). These viruses are mainly transmitted by the fecal-oral route and cause gastroenteritis. However, it can cause other symptoms, which include hemorrhagic conjunctivitis, diabetes (through infection of islet cells), and central nervous system complications [17,18,19,20]. These viruses are acid and heat stable, allowing for their survival in the gastrointestinal tract but inadvertently conferring persistence in WWTPs [21,22]. Infectious EV70 has been detected in the effluents of several WWTPs globally [6,23,24,25]. This indicates that the existing disinfection procedures employed are not adequate to provide safe water for reuse, and there exists a need to explore the efficacy of solar irradiation as a possible additional disinfection strategy against enterovirus 70 (EV70).
In this study, EV70 was exposed to simulated sunlight irradiation for 24 h at a fluence rate of 28.67 J/cm2/h. Aliquots of the virus were harvested at specific time points followed by determination of its infectious titer and RNA concentration. We employed a focus forming assay to overcome the inability of EV70 to replicate well in cell culture [6,26]. To determine if any damage was incurred on the capsid, viruses were assayed for their binding ability to Vero cells. The viral growth kinetics were also assayed by counting the foci generated over a nine-day infection period. Ten days after infection, the genomes of EV70 were sequenced. The assays revealed that irradiated viruses had inhibited replication and binding and harbored nonsynonymous nucleotide substitutions compared to dark-control viruses. Viruses suspended in a wastewater matrix also experienced a significant reduction in viral activity upon exposure to solar irradiation, albeit not as pronounced as that observed when suspended in a saline buffer. Interestingly, all of the irradiated viruses in this study failed to replicate in cell culture, providing a strong endorsement of sunlight as a low-cost natural disinfection strategy.
2. Materials and Methods
2.1. Cells and Viruses
Enterovirus 70 (EV70) was purchased from American Type Culture Collection (ATCC VR-836, Manassas, VA, USA) and propagated in human embryonic kidney (HEK) 293T cells (ATCC CRL-3216). HEK 293T cells were maintained in 75 cm2 flasks (Corning Incorporated, Corning, NY, USA) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1× penicillin and streptomycin (growth medium) (Corning Incorporated, Corning, NY, USA). For the infection study, the HEK 293T cells were seeded in 175 cm2 flasks till confluency and inoculated with EV70 diluted in 10 mL of DMEM supplemented with 2% FBS and 1× penicillin and streptomycin. Flasks were incubated for 1 h at 37 °C and 5% CO2. After incubation, another 10 mL of DMEM supplemented with 2% FBS (virus infection media) was added, and the flasks were returned to the incubator. The cells were observed daily for cytopathic effect (CPE). Once CPE was observed, cells were harvested in the supernatant and pelleted by centrifuging at 2000× g for 10 min. The cells then underwent three freeze-thaw cycles to release any intracellular viruses. The lysate was collected in the supernatant harvested earlier. Next, 30% polyethylene glycol (PEG) 8000 in 0.4 M NaOH was added to the total volume of the lysate and supernatant to a final concentration of 15%. This mixture was stirred at 150 rpm overnight at 4 °C. Viruses were pelleted by centrifuging at 10,000× g for 30 min. The pellet was resuspended in 50 mL of sterile 1× phosphate-buffered saline (PBS) or 0.45 µm-filtered effluent or chlorinated effluent wastewater collected from the Wastewater Treatment Plant of King Abdullah University of Science and Technology (KAUST) [6]. Physical parameters of these collected wastewaters are shown in Table S1. Resuspended viruses were immediately used for solar inactivation.
2.2. Simulated Solar Inactivation Trials
Six milliliters of PEG-purified viruses was dispensed into each of the six microcosms. Each microcosm was made from 5 mL glass beakers (solution depth: 2.7 cm) wrapped in black duct tape to prevent unwanted light penetration. Each microcosm contained a magnetic stirrer and was covered at the top with aluminum foil, in the case of dark controls (n = 3), or left open (n = 3). Each of these microcosms was then placed in 50 mL beakers containing 20 mL of water that served to regulate the temperature at 20 °C. The 50 mL beakers of the dark controls were also covered with aluminum foil, while the irradiated microcosms were covered with a glass filter (Newport Corporation, Irvine, CA, USA) that allowed light of wavelengths ≥280 nm to pass through. Each beaker was then placed in an Atlas Suntest® XLS+ photostimulator (Atlas, Chicago, IL, USA) equipped with a xenon arc lamp. The solar irradiation was measured by a spectroradiometer (ILT950, International Light Technologies, Peabody, MA, USA) to determine the UV irradiance as described previously [27]. The irradiance rate at 280–700 nm was ca. 28 J/cm2/h. The UV irradiance provided by the solar simulator approximated the irradiance measurements of direct noon sunlight measured at two locations on the KAUST campus [27]. Viruses (500 µL) were harvested from the irradiated samples and dark controls after 2 h, 4 h, 6 h, 8 h, 10 h, 12 h, 14 h, 16 h, 20 h, and 24 h (n = 3) of solar inactivation. Virus samples were stored at −80 °C until use.
2.3. Virus Inactivation Kinetics Evaluated by Means of Infectious Assay
Vero cells CCL-81 (American Type Culture Collection, Manassas, VA, USA) were seeded onto 96-well plates at 2 × 104 cells/well in growth media overnight at 37 °C with 5% CO2. Cells were infected with viruses harvested at all time points from the solar inactivation experiment described above. Briefly, viruses were serially diluted (100–10−4), and 50 µL of the diluted inoculum was added to a well with confluent Vero cells for 1 h at 37 °C with 5% CO2. The inoculum was aspirated and replaced with DMEM supplemented with 100 µL of virus infection media. Cells were then returned to the incubator for 16 h. After incubation, cells were washed with 1× sterile PBS and subsequently fixed and permeabilized with 100 µL of ice-cold methanol-acetone (50%-50%) for 10 min. Cells were then washed with 1× PBS three times and incubated with 20 µL of anti-EV antibody (Merck, Cat No. 3321) for 1 h at 37 °C. Cells were then washed with 1× PBS three times and incubated with 20 µL of anti-Mouse (Ms) IgG-FITC (Merck) for 1 h at 37 °C. Cells were washed again with 1× PBS before adding 100 µL of 1× PBS to each of the wells after the last wash. Wells were observed under an epifluorescence microscope for foci (single infected cells). Foci were counted, and the viral titer was estimated using Equation (1):
where FFU/L is focus-forming units per liter and f is the number of fluorescently labeled cells. The infectious titer of irradiated samples was compared against the dark control. Each viral dilution was inoculated in two wells of a 96-well plate, and titration was carried out in triplicate (n = 3). The results from this focus-forming assay were converted to log and natural log (ln) curves by calculating log(Nt/N0) and ln(Nt/N0), respectively, where Nt = virus titer at time t and N0 = virus titer at 0 h (start of experiment). The slopes of the dark-control and irradiated samples (k) were calculated from the ln curves. Prior to the solar inactivation experiments, the absorbance value at 280–700 nm of the PEG-purified virus was determined using a UV-3600 UV–VIS spectrometer (Shimadzu, Kyoto, Japan). The readings were used to generate correction factors that were applied to the slopes of the decay curves prior to half-life calculations and statistical comparison. The half-lives for each experiment, or the durations needed to reduce the viral titer by half, were calculated using the first-order kinetics Equation (2):
where k* is the corrected slope of the inactivation curve and t is time. Statistics were carried out by simple linear regression analysis of the log(Nt/N0) values of dark-control and irradiated samples.
ln(Nt/N0) = −k*t
2.4. Virus Inactivation Kinetics Evaluated by Means of RNA Concentration Decay
RNA was extracted from viruses harvested at each time point throughout the solar inactivation trials by an RNeasy Mini kit (Qiagen, Hilden, Germany). Viral RNA was eluted in 35 µL of water, and the concentration of extracted RNA was determined using the Qubit® single-stranded RNA assay kit with the Qubit® fluorometer (Thermo Fisher Scientific, Carlsbad, CA, USA). Decay kinetics and statistical comparisons were carried out similarly as described in the previous section. Specifically, the RNA concentration obtained at the start of the experiment was defined as N0 and was expressed in log(Nt/N0) and ln(Nt/N0) equations, where Nt is the RNA concentration of the virus sample harvested at time t and N0 is the RNA concentration harvested from 0 h (start of experiment).
2.5. Growth Curve Analysis
Vero cells were seeded onto 96-well plates at a cell density of 2 × 104 cells per well in growth media overnight at 37 °C with 5% CO2. Cells were then inoculated with 50 µL of serial dilutions (100–10−4) of T0, D24 or L24 at the multiplicity of infection of 0.1, where T0 is presolar-irradiated EV70 (wild-type), D24 is the dark-control EV70 post solar irradiation, and L24 is EV70 that had undergone 24 h of simulated solar irradiation. Cells were incubated at 37 °C with 5% CO2 for 1 h for viral absorption. Each dilution was added to two wells of a 96-well plate, and the titration for each virus sample was done in duplicate (n = 2). Virus inoculum was removed, replaced with viral infection medium, and incubated for infection to take place. Cells were fixed with ice cold methanol-acetone (1:1 v/v) for 10 min at 1, 3, 5, 7, and 9 days post-infection (dpi). Cells were subsequently labeled with anti-EV and anti-Ms IgG-FITC antibodies, and the corresponding viral titers were calculated as described above.
2.6. Virus Absorption Assay
Vero cells were seeded in 6-well plates at a cell density of 1.2 × 106 cells per well in growth media overnight at 37 °C with 5% CO2. Cells were then inoculated in 100 µL of T0, D24 or L24 diluted in 500 µL of 1× sterile PBS for 1 h at 4 °C. After binding, cells were washed with 1× sterile PBS and subsequently scraped and collected in 400 µL of 1× sterile PBS. This cell suspension went through three rounds of freeze-thaw to release bound viruses and were then serially diluted (100–10−4) and titered in a similar manner as described in Section 2.3.
2.7. EV70 Genome Sequencing
Vero cells were seeded in the same conditions as for the viral absorption assay described in Section 2.6. Cells were then inoculated with similar titers of T0, D24 or L24 in 500 µL of 1× sterile PBS for 1 h at 37 °C with 5% CO2. The viral inoculum was replaced with DMEM supplemented with 2% FBS and 1× penicillin and streptomycin. Cells were placed in the incubator for 10 days. At 10 days post-infection (dpi), cells were washed, scraped and collected in 400 µL of 1× sterile PBS. RNA was extracted from the cells by the RNeasy Mini kit (Qiagen, Hilden, Germany). RNA was used as the template for fragment Polymerase Chain Reaction (PCR) where the genome was amplified into 18 overlapping fragments of 750 base pairs. Reaction mixes were prepared by adding 25 µL of 2× RT Buffer, 1.5 µL of forward and reverse primers at 10 µM concentration, 1 µL of Life Technologies SuperScriptTM II RT enzyme (Thermo Fisher Scientific, Carlsbad, CA, USA), 18 µL of water and 0.83 µL of template RNA. Primers for each of the fragments are listed in Table S2.
Fragments were amplified by touchdown PCR, which included cDNA synthesis at 55 °C, for 30 min, an initial denaturation step of 94 °C, for 2 min, followed by 15 cycles of 94 °C for 15 s, annealing at 52 °C for 30 s, and extension at 68 °C for 100 s, with the annealing temperature decreasing by 1 °C with each cycle. This was followed by another 30 cycles of 94 °C for 30 s, annealing at 48 °C for 30 s, and extension at 68 °C for 100 s. A final extension at 72 °C for 5 min was performed. Amplicons were run on a 1.2% agarose gel and visualized by SYBR Green (Thermo Fisher Scientific, Carlsbad, CA, USA). Bands corresponding to ~750 bp were extracted using the Wizard® SV Gel and PCR Clean-Up system (Promega, Fitchburg, WI, USA). Purified PCR products were sent to the KAUST Genomics Core lab for Sanger sequencing. PCR sequences were aligned using the SeqMan program of the DNASTAR’s Lasergene software package (DNASTAR, Madison, WI, USA). Aligned contigs were saved as consensus sequences for each of the viral samples. Consensus sequences were aligned in BioEdit Sequence Alignment Editor [28] and translated in silico. The amino acid sequences of each generated genome were submitted to the Phyre2 web Portal for 3D structure prediction [29]. The .pdb files generated from Phyre2 were visualized in PyMOL [30].
3. Results
3.1. Viral Inactivation Upon Solar Irradiation
The infectious capacities of irradiated EV70 in phosphate-buffered saline (PBS), as well as in effluent and in chlorinated effluent wastewater matrices were assayed against dark-control EV70 (Figure 1A). After 24 h of irradiation, at a fluence of 688 J/cm2, the infectivity of dark-control EV70 was reduced by 0.18 ± 0.07 log, 0.11 ± 0.003 log and 0.12 ± 0.03 log in PBS, effluent and chlorinated effluent matrices, respectively (kobs = 0.014, 0.010 and 0.009) (Figure 1A and Table 1A). One-way ANOVA revealed no significant difference between the decay constants of the dark controls in the three different matrices (p > 0.1). Linear regression analysis of the slopes of the dark control showed no positive correlation with respect to time, suggesting that dark-control viruses were relatively stable in each of the matrices over a 24 h period (p > 0.01).
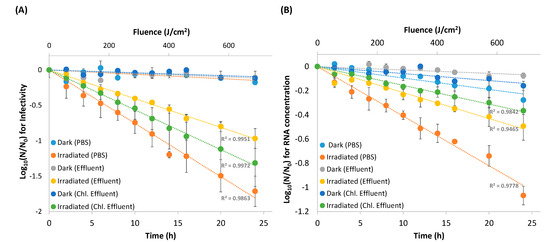
Figure 1.
Decay curves of EV70 under simulated solar irradiation (●, ●, ●) compared to dark-control EV70 (●, ●, ●) over 24 h for viruses suspended in phosphate-buffered saline (PBS) (n = 3), effluent matrix (n = 2), and chlorinated effluent matrix (n = 2). The irradiance rate at 280–700 nm was 27.86 J/cm2/h. (A) Viruses were harvested at each time point and subsequently tittered. (B) RNA was extracted from the viruses harvested at each time point and subsequently quantified.

Table 1.
Decay kinetic constants and half-life of (A) EV70 infectivity, and (B) RNA concentration, under simulated solar irradiation for 24 h in phosphate-buffered saline (PBS) (n = 3), effluent wastewater matrix (n = 2), and chlorinated effluent wastewater matrix (n = 2). kobs = decay constant. t1/2 = half-life of decay.
In contrast, irradiated EV70 reduced in infectivity by 1.7 ± 0.2, 1.0 ± 0.1, and 1.3 ± 0.3-logs in PBS, effluent and chlorinated effluent with decay constants of 1.4, 0.9 and 1.0 (Figure 1A and Table 1A). t-test analysis between the decay constants of the dark-control and irradiated samples showed that the decay within each matrix was significant (p < 0.05). One-way ANOVA of the decay constants of the irradiated samples revealed that the decay in each of the matrices was significantly different from the others, with the decay in PBS being the fastest (t1/2 = 30 ± 3 min), followed by the decay in chlorinated effluent (t1/2 = 41 ± 3 min), then effluent (t1/2 = 47 ± 5 min) (p < 0.05) (Figure 1A and Table 1A).
3.2. RNA Decay
After the same dose of simulated solar irradiation, the RNA concentration from dark-control EV70 decreased by 0.3 ± 0.1 log, 0.1 ± 0.02 log, and 0.2 ± 0.04 log in PBS, effluent and chlorinated effluent matrices, respectively (kobs = 0.002, 0.007, and 0.013). One-way ANOVA revealed that the decay constants of the dark control did not differ between the three matrices (p > 0.05) (Figure 1B and Table 1B). Linear regression analysis of the dark control showed a positive correlation with respect to time, suggesting that RNA of the dark-control samples was not stable in any matrix (p < 0.05).
The RNA concentrations of the irradiated samples decayed by 1.1 ± 0.1 log, 0.5 ± 0.1 log, and 0.4 ± 0.1 logs in PBS, effluent and chlorinated effluent, respectively (kobs = 0.77, 0.48, and 0.29) (Figure 1B and Table 1B). Within each matrix, the decay constant of the irradiated samples differed significantly from the dark control, suggesting that simulated solar irradiation sped up RNA decay in EV70 (p < 0.05). The decay constants of the irradiated samples did not differ significantly from each other (p > 0.05), but RNA from EV70 in PBS decayed the fastest (t1/2 = 57 ± 14 min), followed by EV70 in effluent wastewater matrix (t1/2 = 88 ± 9 min) and then EV70 in chlorinated effluent wastewater matrix (t1/2 = 145 ± 7 min) (Table 1B).
3.3. Irradiated EV70 Displays Inhibited Viral Replication
To study the replication kinetics of irradiated viruses, Vero cells were infected with the same titer of T0, D24 or L24 for nine days. Focus-forming units were counted throughout this period to produce the growth curves presented in Figure 2. Sixteen hours post-infection (hpi), T0 in PBS replicated to 3.9 × 103 ± 8.1 × 102 FFU/mL, and D24 in PBS replicated to 4.7 × 103 ± 1.1 × 103 FFU/mL. Both T0 and D24 peaked on the fifth day post-infection (dpi) at titers of 7.7 × 104 ± 4.1 × 103 FFU/mL and 6.3 × 104 ± 1.0 × 104 FFU/mL, respectively. A titer of 2.9 × 104 ± 1.0 × 104 FFU/mL and 3.2 × 104 ± 1.4 × 104 FFU/mL was observed for both these viruses at 9 dpi, respectively. This apparent reduction in titer was probably due to the detachment of infected cells from the monolayer nine days after infection. In contrast, L24 in PBS replicated to 6.4 × 103 ± 1.3 × 103 FFU/mL 16 hpi and remained relatively similar over the 9-day period. Nor did L24 display a peak at 5 dpi, as seen in T0 and D24 in PBS (Figure 2A).
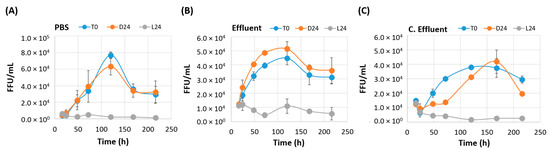
Figure 2.
Growth kinetics of nontreated EV70 (T0 ●), dark-control EV70 (D24 ●), and simulated-solar-irradiated EV70 (L24 ●). Confluent monolayers were infected with similar concentrations of each of the viral samples, and the foci formed over a nine-day period were enumerated. (A) EV70 resuspended in phosphate-buffered saline (PBS) (n = 3). (B) EV70 resuspended in effluent wastewater matrix (n = 2). (C) EV70 resuspended in chlorinated effluent wastewater matrix (n = 2).
T0 in effluent matrix replicated to 1.1 × 104 ± 2.6 × 102 FFU/mL at 16 hpi and peaked to 4.5 × 104 ± 4.7 × 103 FFU/mL at 5 dpi, and this titer reduced to 3.1 × 104 ± 4.5 × 103 FFU/mL at 9 dpi. D24 in effluent matrix exhibited a similar growth pattern, with a titer of 1.2 × 104 ± 1.4 × 103 FFU/mL at 16 hpi, 5.1 × 104 ± 5.2 × 103 FFU/mL at 5 dpi and 3.6 × 104 ± 9.1 × 103 FFU/mL at 9 dpi. L24 in effluent matrix replicated to 1.1 × 104 ± 1.8 × 103 FFU/mL at 16 hpi. On 5 dpi, when T0 and D24 replicated to peak titers, L24 in effluent matrix only displayed a titer of 1.1 × 104 ± 5.1 × 103 FFU/mL. The titer of L24 in effluent matrix did not exceed the titer displayed at 16 hpi throughout the course of the experiment (Figure 2B).
In the chlorinated effluent wastewater matrix, T0 replicated to 1.5 × 104 ± 1.4 × 102 FFU/mL at 16 hpi and peaked at approximately 4.0 × 104 FFU/mL approximately 130 hpi. It reached 3.0 × 104 ± 2.5 × 103 FFU/mL at 9 dpi. D24 in chlorinated effluent replicated to 1.3 × 104 ± 1.3 × 103 FFU/mL at 16 hpi and peaked at 4.2 × 104 ± 8.1 × 103 FFU/mL at 7 dpi, before finally reaching a titer of 2.0 × 104 at 9 dpi. In contrast, L24 in chlorinated effluent replicated to 1.3 × 104 ± 1.6 × 103 FFU/mL at 16 hpi and decreased to 2.6 × 103 ± 9 × 101 FFU/mL by 9 dpi (Figure 2C). T0 and D24 in chlorinated effluent peaked later than in PBS or effluent, possibly due to the presence of residual disinfection byproducts that may have a toxic effect on mammalian cell lines [31].
3.4. Irradiated EV70 Displayed Reduced Binding Capability
Untreated EV70 (T0) and dark-control EV70 harvested at 24 h post-irradiation (D24) displayed similar binding affinities to Vero cells in cell culture in all three matrices (Figure 3) (p > 0.05). EV70 in PBS displayed the greatest reduction in binding among the three matrices, 2.6 ± 0.3 log, followed by EV70 in effluent matrix at 1.8 ± 0.3 log and EV70 in chlorinated effluent matrix at 1.3 ± 0.2 log. One-way analysis of variance (ANOVA) revealed that the log reduction values (LRV) of the irradiated samples were significantly different from each other (p < 0.01), suggesting that the matrix affected the binding affinity of EV70 to Vero cells in cell culture (Figure 3).
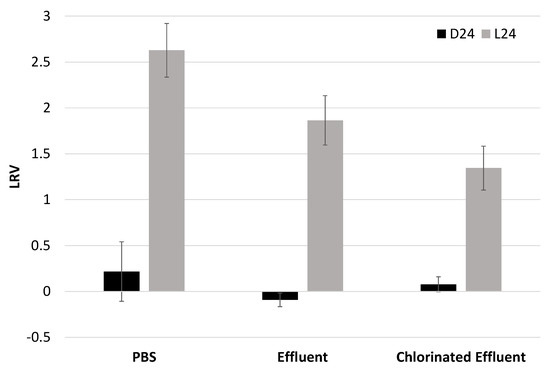
Figure 3.
Log reduction values of the binding affinity of dark-control viruses (D24) and solar-irradiated EV70 (L24) with respect to untreated EV70. Y-axis represents log reduction value (LRV) with respect to nontreated EV70. D24: dark-control EV70 viruses that were placed in the solar simulator for 24 h but kept in the dark. L24: EV70 viruses that were exposed to simulated solar irradiation for 24 h. The irradiance rate at 280–700 nm was 28 J/cm2/h. PBS, n = 3; effluent, n = 2; chlorinated effluent, n = 2.
3.5. Irradiated Viruses Select for Five Nonsynonymous Mutations
D24 and L24 from all three water matrices were infected in Vero cells for 10 dpi. Viral RNA was extracted and amplified into 18 overlapping fragments for Sanger-based sequencing. Figure 4 shows the alignment of the nucleotide sequence and the in silico-translated amino acid sequence. Nucleotide numbers here start at the first ATG of the coding sequence of the reference strain EV70 J670/71 (GenBank D00820.1) [32]. D24 in all matrices and in both replicates in PBS yielded the same nucleotide sequence, showing that any sequence difference seen in L24 was an effect of solar irradiation. L24 in PBS displayed five nonsynonymous nucleotide substitutions (Figure 4). A40G and C809T were observed in the VP4 and VP2 genes, respectively. These mutations resulted in conserved-amino-acid substitutions: Lys14Glu and Ala201Val (Figure 4A,B). Both these changes occurred in unstructured motifs of their respective proteins (Figure S1). G1171A was observed in the VP3 gene, which caused a nonconserved-amino-acid substitution of Gly71Ser (Figure 4C). However, this mutation maintained the β-sheet structure of this protein (Figure S1). G1810C was observed in the VP1 gene, resulting in Glu50Gln substitution, which occurred in an unstructured region of the protein (Figure 4D and Figure S1). A4801C in the 3Cpro gene resulted in the conserved-site substitution of Ile47Leu (Figure 4E). No structural changes were observed due to this mutation (Figure S1). Out of these five nonsynonymous mutations, A40G was also seen in L24 in chlorinated effluent matrix (Figure 4A). L24 in both wastewater matrices harbored a synonymous mutation of G4698A in the 3Cpro gene, which was not observed in L24 in PBS (Figure 4F).
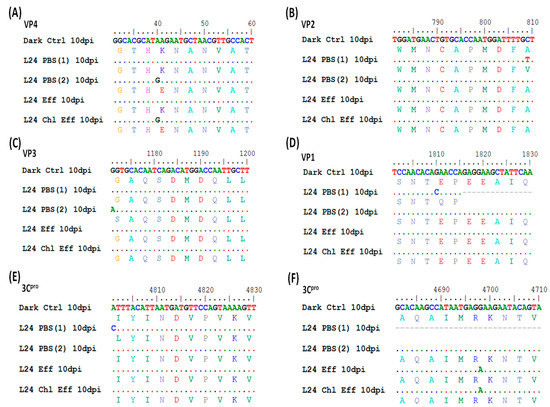
Figure 4.
Genome sequence analysis of D24 and L24 (in PBS, effluent and chlorinated effluent) 10 days post-infection (dpi) in Vero cells. The nucleotide sequence of D24 in PBS is taken as the reference strain in this alignment (top row). The second (and other even-numbered) rows represent the predicted amino acid sequence. Identical nucleotide sequences are represented by a dot (.). Six nonsynonymous mutations were observed in the genes coding for (A) VP4, (B) VP2, (C) VP3, (D) VP1, and (E,F) 3Cpro. The number of experiments performed was n = 2 for EV70 in PBS, n = 1 in effluent (Eff) and n = 1 in chlorinated effluent (Chl Eff). Sequence data could only be obtained from one experimental run for solar inactivation in wastewater matrices. Scale (top row) represents the nucleotide sequence of EV70 strain J670/71 (from NCBI reference D00820), with position 1 corresponding to the first nucleotide of the open reading frame.
4. Discussion
Earlier observations of viable and infective viruses in post-treated effluent provided the main impetus for this study [6], as their presence can complicate the reuse of reclaimed waters. To circumvent viral risks, chlorine disinfection is typically performed at the last step of a wastewater treatment process. However, chlorine works with varying effectiveness against different types of viruses [5]. This led to the suggestion of including combinations of various disinfection processes in a single WWTP. However, retrofitting different modular units of disinfection processes may incur additional operating costs. Solar disinfection of treated wastewater was therefore studied to provide a natural, low-cost and abundant disinfection strategy to further inactivate remnant viruses present in the reclaimed waters.
Specifically, EV70 was chosen as a model organism in this study, as infectious enteroviruses were previously found after wastewater treatment in concentrations approximating the infectious dose [6,33]. EV70 has not been studied extensively for its susceptibility to disinfectants due to its lack of plaque-producing capability in cell culture. To overcome this hurdle, a focus-forming assay was employed, which measured viral titer by fluorescently labeling virus-infected cells with virus-specific antibodies. This technique also required shorter duration compared to a traditional plaque assay [26].
We observed that EV70 in PBS experienced a 1.7-log reduction in infectivity after a dose of 688 J/cm2 (Figure 1). This is consistent with the finding that poliovirus type 2 experienced a 4-log reduction with a simulated solar irradiation of 1224 J/cm2, which is equivalent to a 2-log reduction at approximately 612 J/cm2 [14]. Both EV70 and polioviruses are from the Picornaviridae family and have similar sizes (approximately 30 nm in diameter), capsid structures and genome lengths (EV70: 7200 nt, poliovirus: 7500 nt) [34,35]. In contrast, other members of Picornaviridae require differing doses of solar irradiation to achieve a similar reduction in infectivity. For example, Coxsackie viruses require approximately 58.5–99 J/cm2, and ECHO viruses require 50–60 J/cm2 of solar irradiation to achieve a 2-log reduction [15,16]. Both Coxsackie and ECHO viruses have similar sizes (28 nm and 24–30 nm, respectively) and genome lengths (approximately 7400 nt and 7500 nt, respectively) to EV70 [36,37,38]. The data presented in this study agree with earlier studies that infer the need for varying solar fluence to inactivate different viral species. While the structures of viruses are generally similar within a family, species might differ in protein folding and genome secondary structure, which give rise to differences in susceptibility to solar irradiation [39].
Picornaviridae have a positively stranded RNA genome that is directly translated by host-cell ribosomes [13]. Here, damage to the genome was indicated by the decay in the RNA concentrations in the presence of solar irradiation (Figure 1B). In addition, the reduction in binding capacity of L24 indicated conformational damage to the capsid, stopping it from recognizing the viral receptor on the Vero cells (Figure 3). This reduction in binding was of a larger magnitude than the reduction in infectivity as seen in Figure 1A at 24 h across all three matrices. Since receptor binding is the first step in a virus replication cycle, any irradiation-induced damage to the capsid could result in the inability of the capsid to recognize the receptor on host cells. Hence, this decrease in binding of L24 was most likely the primary cause for the decrease in infectious capacity.
Although EV70 with a damaged capsid may have a reduced binding capability, replication would theoretically still be possible if the interior structure of the viral particle remained undamaged. To test this, we performed a growth curve analysis, which showed that L24 was unable to replicate to similar titers as T0 or D24 even after 9 dpi in all three water matrices (Figure 2). This was observed despite the similar multiplicity of infection between T0, D24, and L24. This information indicates an inability of solar-irradiated EV70 to replicate as effectively as wild-type or dark-control viruses. The capsids of picornaviruses undergo a dramatic antigenic alteration before the virus uncoats [40]. Translation is then initiated by the internal ribosomal entry sites in the 5′ untranslated region of the genome, which is composed of five stem-loops (II-VI) [41,42]. Viral translation is also promoted by the binding of host-cell IRES trans-acting factors, such as FBP1-3, hnRNP K and hnRNP A, which recognize the 5′ untranslated region of the viral genome [42,43,44,45]. Solar irradiation could induce structural damage to the capsid and genome of EV70, leading to reduced binding and replication capacity. Not only would a structurally damaged capsid fail to bind to the host-cell receptor, but it might fail to undergo the antigenic alteration necessary for uncoating to occur [40]. UV irradiation promotes RNA-protein cross-linking [46]. The formation of covalent bonds between the EV70 genome and the capsid might affect the release of the RNA out of the capsid during the uncoating process. The integrity of the cloverleaf and stem loop structure present in the 5’ untranslated region of the EV70 genome might be negatively affected by solar irradiation. A disintegration of structure in this region of the genome may prevent successful docking of the ribosome and other host-cell translation initiation factors. Lastly, owing to the structural damage to the genome, translation might not proceed as efficiently as in wild-type viruses, producing proteins which might not support viral replication.
To elucidate if mutations did indeed occur in key proteins of EV70, we sequenced the coding region of the genome. Initially, fragment PCR of viruses directly sampled after 24 h of solar irradiation was performed (data not shown). However, this did not yield sufficient concentrations of PCR amplicons for sequencing. To overcome this technical constraint, Vero cells were infected with L24 or D24 for 10 days, and the viral RNA, which had amplified in the course of the infection, was extracted and sequenced. The sequence of the L24 viral genome derived from this experiment is, hence, not a direct product of solar irradiation but was selected for 10 dpi. This genome could be viewed as an ‘escape mutant’, being the only sequence that had replicated enough to be amplified by PCR. However, this sequence was still unable to replicate as effectively as T0 or D24 (Figure 2).
The irreproducibility of nucleotide substitutions between trials 1 and 2 for L24 in PBS indicate that solar irradiation induces mutations in a random manner. However, four out of the six mutations listed occurred in the capsid genes, which are at the 5′ end of the genome. Positions 40 and 4801 also showed mutations in two of the four irradiated samples (Figure 4A,F). These findings might suggest that the capsid genes, as well as position 4801 in the 3Cpro gene, are more prone to mutation by solar irradiation compared to the rest of the genome.
The structure of the capsid of bovine enterovirus (BEV), a picornavirus, has been determined [47]. The structural proteins of BEV share 48% identity with EV70 [48], and its tertiary structure is collinear with other enteroviruses [49]. Comparisons with the amino acid sequence of BEV’s capsid reveal that the amino acid substitutions of EV70 listed in this study did not occur in any of the known functional motifs. However, an earlier study showed that an introduction of a single amino acid substitution at five different positions in the capsid genes resulted in a change in viral tropism [50]. These proteins constitute the capsid and form the depression known as the ‘canyon’, which recognizes the cellular receptor DAF/CD55 for attachment to the host [51,52]. VP1, which is the most exposed protein of the capsid of Picornavirus [53], forms a hydrophobic pocket that allows for myristic acid binding [54] and is believed to be involved in the binding of metal ions [55,56]. VP1 is also believed to have a role in the uncoating of the virus particle [50]. Even though Glu50Gln in VP1 occurred in an unstructured motif (Figure S1), the substitution might alter the charge of VP1.
EV70 with a glutamic acid instead of a lysine at position 14 of VP4 protein replicates poorly in HeLa cells [50]. In this current study, L24 in PBS and chlorinated effluent displayed this substitution (Figure 4), which may have accounted for the poor replication. It is likely that this mutation resulted in a change in the charge of the overall protein, as lysine is typically positive at neutral pH while glutamic acid is negatively charged. This would have resulted in poor binding of EV70 to the host cells. Similarly, even though the amino acid substitution Gly71Ser did not affect the folding of the β-sheet in VP3 (Figure S1), the overall polarity of the protein might have been affected owing to the polar nature of serine as opposed to glycine. Both these substitutions might have synergistically affected viral function.
In addition to assessing changes in the capsid proteins, the 3Cpro protein of Picornaviridae was also assessed since this protein displays a multitude of functions in the infected cell. Initially shown to be a protease that cleaves the functional proteins from the polyprotein precursor, 3Cpro also cleaves host-cell proteins to shut down host-cell transcription, translation, and nucleo-cytoplasmic trafficking and promote apoptosis (reviewed in [57]). There exist four main functional domains in the 3Cpro protein: the N-terminal domain (aa 12–13), the central domain (aa 82–86), the β-ribbon (aa 123–133) and the C-terminal domain (aa 154–156) [58,59,60]. The amino acid substitution Ile47Leu occurred in between the N-terminal domain and the central domain, an area that lacks any known function (Figure 4). It is important to note that this substitution did not affect the integrity of the β-sheet motif of this protein (Figure S1). However, further investigations into this amino acid substitution should be carried out to determine if the function of the 3Cpro protein is altered. If this substitution results in a change in the function of the protein, this could explain the reduced ability of L24 to replicate to high titers, as seen in cells infected with T0 and D24 (Figure 2). This could be a result of inadequate cleavage of the viral polyprotein or inadequate suppression of host-cell factors, allowing for the host cell to overcome the viral replication machinery.
In addition to observing a significant impact on the viral infectivity and persistence due to solar irradiation, we also observed that viral inactivation occurred at a slower rate when the viruses were present in wastewater matrix. This concurs with earlier observations [61,62,63]. Furthermore, out of the six nucleotide mutations found, only 2 were seen in L24 in wastewater matrices, A40G and G4698A, while L24 in PBS had 5 mutations (Figure 4). This indicates that viruses in the wastewater are less susceptible to UV-B [12,16]. Effluent and chlorinated effluent wastewaters used in this study had a total organic carbon (TOC) concentration of 4.2 mg/L and 5.2 mg/L, respectively, while PBS had undetected levels of TOC, as expected (Table S1). These organic compounds can act as radical scavengers [64,65], reduce light intensity [66], or encapsulate viruses with a protective organic coating that makes them more resistant to external environmental stressors when present in wastewaters. The latter has been alluded to by the findings that non-enveloped viruses are stable in wastewaters [67,68]. Alternatively, the high alkalinity in wastewater might favor the reaction between bicarbonates and hydroxyl radicals formed upon solar irradiation. This reaction results in the generation of CO3●- which reacts slower with organic molecules compared to ●O2 radicals [27,69,70].
While these reasons could explain the slower inactivation rates of EV70 in wastewater matrices, it is important to note that irradiated viruses, irrespective of matrix, all failed to propagate in cell culture (Figure 4). This indicates that solar irradiation successfully inhibits viral replication in cell culture, preventing the generation of infectious viral progeny in all three water matrices evaluated in this study. This strongly suggests that solar irradiation modifies the replication capacity of EV70 to the point that it might not pose a significant public health threat. Although the data from this study suggest that solar irradiation may serve as a good disinfection technique, its efficacy may be lower in turbid waters due to lower solar penetration and higher light-scattering effect. Operators would also need to create a holding tank that is shallow enough to allow for proper solar penetration and irradiation. This would not be feasible in densely populated places with limited land space. Hence, the use of solar irradiation as an effective, natural, and low-cost disinfection strategy against EV70 would only be feasible for use in low-turbidity waters, presumably in permeates after membrane filtration processes, and in places unconstrained by land availability.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4441/11/1/64/s1, Figure S1: Predicted structure of EV70, Table S1: Physical parameters of the matrices used in this study, Table S2: Sequences and names of primers used in this study.
Author Contributions
M.R.J. and P.-Y.H. conceived and planned the experiments. M.R.J. carried out the experiments and performed the analysis. M.R.J. and P.-Y.H. wrote the manuscript together. P.-Y.H. provided reagents and materials. All authors have read and approved the manuscript.
Funding
This research was supported by the KAUST baseline funding BAS/1/1033/-01-01 awarded to P.-Y.H.
Acknowledgments
The authors would like to thank George Princeton Dunsford for access to the KAUST wastewater treatment plant, Moustapha Harb for providing sampling assistance, Nada Al Jassim for training required to operate the solar simulator and Noor Zaouri for assistance in the LC-OCD analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- AghaKouchak, A.; Feldman, D.; Hoerling, M.; Huxman, T.; Lund, J. Water and climate: Recognize anthropogenic drought. Nature 2015, 524, 409–411. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Da, L.; Song, K.; Li, B.-L. Temporal variations of surface water quality in urban, suburban and rural areas during rapid urbanization in shanghai, china. Environ. Pollut. 2008, 152, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, C.; Yamada, K. Impact of population growth on the water quality of natural water bodies. Sustainability 2017, 9, 1405. [Google Scholar] [CrossRef]
- Scholz, M. Chapter 19—Disinfection. In Wetlands for Water Pollution Control, 2nd ed.; Scholz, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 129–136. [Google Scholar]
- Zhong, Q.; Carratalà, A.; Ossola, R.; Bachmann, V.; Kohn, T. Cross-resistance of uv- or chlorine dioxide-resistant echovirus 11 to other disinfectants. Front. Microbiol. 2017, 8, 1928. [Google Scholar] [CrossRef]
- Jumat, M.; Hasan, N.; Subramanian, P.; Heberling, C.; Colwell, R.; Hong, P.-Y. Membrane bioreactor-based wastewater treatment plant in saudi arabia: Reduction of viral diversity, load, and infectious capacity. Water 2017, 9, 534. [Google Scholar] [CrossRef]
- Lo, H.-L.; Nakajima, S.; Ma, L.; Walter, B.; Yasui, A.; Ethell, D.W.; Owen, L.B. Differential biologic effects of cpd and 6-4pp uv-induced DNA damage on the induction of apoptosis and cell-cycle arrest. BMC Cancer 2005, 5, 135. [Google Scholar] [CrossRef]
- Drouin, R.; Therrien, J.P. Uvb-induced cyclobutane pyrimidine dimer frequency correlates with skin cancer mutational hotspots in p53. Photochem. Photobiol. 1997, 66, 719–726. [Google Scholar] [CrossRef]
- Lamont, Y.; Rzeżutka, A.; Anderson, J.; MacGregor, S.; Given, M.; Deppe, C.; Cook, N. Pulsed uv-light inactivation of poliovirus and adenovirus. Lett. Appl. Microbiol. 2007, 45, 564–567. [Google Scholar] [CrossRef]
- Lizasoain, A.; Tort, L.F.L.; García, M.; Gillman, L.; Alberti, A.; Leite, J.P.G.; Miagostovich, M.P.; Pou, S.A.; Cagiao, A.; Razsap, A.; et al. Human enteric viruses in a wastewater treatment plant: Evaluation of activated sludge combined with uv disinfection process reveals different removal performances for viruses with different features. Lett. Appl. Microbiol. 2018, 66, 215–221. [Google Scholar] [CrossRef]
- Chevrefils, G.; Caron, É.; Wright, H.; Sakamoto, G.; Payment, P.; Barbeau, B.; Cairns, B. Uv dose required to achieve incremental log inactivation of bacteria, protozoa and viruses. IUVA News 2006, 8, 38–45. [Google Scholar]
- Lytle, C.D.; Sagripanti, J.L. Predicted inactivation of viruses of relevance to biodefense by solar radiation. J. Virol. 2005, 79, 14244–14252. [Google Scholar] [CrossRef] [PubMed]
- Racaniello, V.R. Picornaviridae: The viruses and their replication. In Fields virology, sixth edition; Wolters Klewer/Lippincott Williams Wilkinspp: Philadelphia, PA, USA, 2013; Volume 1, pp. 453–489. [Google Scholar]
- Heaselgrave, W.; Patel, N.; Kilvington, S.; Kehoe, S.; McGuigan, K. Solar disinfection of poliovirus and acanthamoeba polyphaga cysts in water—A laboratory study using simulated sunlight. Lett. Appl. Microbiol. 2006, 43, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Heaselgrave, W.; Kilvington, S. The efficacy of simulated solar disinfection (sodis) against coxsackievirus, poliovirus and hepatitis a virus. J. Water Health 2012, 10, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Gerba, C.P.; Gramos, D.M.; Nwachuku, N. Comparative inactivation of enteroviruses and adenovirus 2 by uv light. Appl. Environ. Microbiol. 2002, 68, 5167–5169. [Google Scholar] [CrossRef] [PubMed]
- Mirkovic, R.R.; Kono, R.; Yin-Murphy, M.; Sohier, R.; Schmidt, N.J.; Melnick, J.L. Enterovirus type 70: The etiologic agent of pandemic acute haemorrhagic conjunctivitis. Bull. World Health Organ. 1973, 49, 341–346. [Google Scholar]
- Khetsuriani, N.; Lamonte-Fowlkes, A.; Oberst, S.; Pallansch, M.A. Enterovirus surveillance—United States, 1970–2005. MMWR Surveill Summ 2006, 55, 1–20. [Google Scholar] [PubMed]
- Roivainen, M. Enteroviruses: New findings on the role of enteroviruses in type 1 diabetes. Int. J. Biochem. Cell Biol. 2006, 38, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Ylipaasto, P.; Klingel, K.; Lindberg, A.M.; Otonkoski, T.; Kandolf, R.; Hovi, T.; Roivainen, M. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia 2004, 47, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Blomqvist, S.; Savolainen, C.; Raman, L.; Roivainen, M.; Hovi, T. Humanrhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J. Clin. Microbiol. 2002, 40, 4218–4223. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Li, D.; Xie, G.; Hu, Y.; Zhang, Q.; Kong, X.; Guo, N.; Li, Y.; Duan, Z. Inactivation of ev71 by exposure to heat and ultraviolet light. Bing Du Xue Bao 2015, 31, 500–506. [Google Scholar] [PubMed]
- Ottoson, J.; Hansen, A.; Westrell, T.; Johansen, K.; Norder, H.; Stenstrom, T.A. Removal of noro- and enteroviruses, giardia cysts, cryptosporidium oocysts, and fecal indicators at four secondary wastewater treatment plants in sweden. Water Environ. Res. 2006, 78, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Lee, B.E.; Neumann, N.; Ashbolt, N.; Craik, S.; Maal-Bared, R.; Pang, X.L. Assessment of human virus removal during municipal wastewater treatment in edmonton, canada. J. Appl. Microbiol. 2015, 119, 1729–1739. [Google Scholar] [CrossRef] [PubMed]
- Simmons, F.J.; Xagoraraki, I. Release of infectious human enteric viruses by full-scale wastewater utilities. Water Res. 2011, 45, 3590–3598. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Gastaminza, P.; Cheng, G.; Kapadia, S.; Kato, T.; Burton, D.R.; Wieland, S.F.; Uprichard, S.L.; Wakita, T.; Chisari, F.V. Robust hepatitis c virus infection in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 9294–9299. [Google Scholar] [CrossRef] [PubMed]
- Al-Jassim, N.; Mantilla-Calderon, D.; Wang, T.; Hong, P.-Y. Inactivation and gene expression of a virulent wastewater escherichia coli strain and the nonvirulent commensal escherichia coli dsm1103 strain upon solar irradiation. Environ. Sci. Technol. 2017, 51, 3649–3659. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Sympos. Ser. 1999, 41, 95–98. [Google Scholar]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845. [Google Scholar] [CrossRef]
- DeLano, W.L.; Lam, J.W. Pymol: A communications tool for computational models. Abstr. ACS 2005, 230, U1371–U1372. [Google Scholar]
- Wagner, E.D.; Plewa, M.J. Cho cell cytotoxicity and genotoxicity analyses of disinfection by-products: An updated review. J. Environ. Sci. 2017, 58, 64–76. [Google Scholar] [CrossRef]
- Ryan, M.D.; Jenkins, O.; Hughes, P.J.; Brown, A.; Knowles, N.J.; Booth, D.; Minor, P.D.; Almond, J.W. The complete nucleotide sequence of enterovirus type 70: Relationships with other members of the picornaviridae. J. Gen. Virol. 1990, 71, 2291–2299. [Google Scholar] [CrossRef] [PubMed]
- Cliver, D.O. Experimental infection by waterborne enteroviruses. J. Food Prot. 1981, 44, 861–865. [Google Scholar] [CrossRef]
- Hogle, J.M. Poliovirus cell entry: Common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 2002, 56, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.R.; McCormick, M.J. Electron microscopy of poliomyelitis virus. Yale J. Biol. Med. 1956, 28, 589–597. [Google Scholar] [PubMed]
- Chen, L.; Yang, H.; Feng, Q.-J.; Yao, X.-J.; Zhang, H.-L.; Zhang, R.-L.; He, Y.-Q. Complete genome sequence of a coxsackievirus a16 strain, isolated from a fatal case in Shenzhen, Southern China, in 2014. Gen. Ann. 2015, 3, e00391-15. [Google Scholar] [CrossRef] [PubMed]
- Crowell, R.L.; Landau, B.J. A short history and introductory b. In The Coxsackie B Viruses; Tracy, S., Chapman, N.M., Mahy, B.W.J., Eds.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 1–11. [Google Scholar]
- Dahllund, L.; Nissinen, L.; Pulli, T.; Hyttinen, V.P.; Stanway, G.; Hyypia, T. The genome of echovirus 11. Virus Res. 1995, 35, 215–222. [Google Scholar] [CrossRef]
- Silverman, A.I.; Peterson, B.M.; Boehm, A.B.; McNeill, K.; Nelson, K.L. Sunlight inactivation of human viruses and bacteriophages in coastal waters containing natural photosensitizers. Environ. Sci. Technol. 2013, 47, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Fenwick, M.L.; Wall, M.J. Factors determining the site of synthesis of poliovirus proteins: The early attachment of virus particles to endoplasmic membranes. J. Cell Sci. 1973, 13, 403–413. [Google Scholar] [PubMed]
- Fitzgerald, K.D.; Semler, B.L. Bridging ires elements in mrnas to the eukaryotic translation apparatus. Biochim. Biophys. Acta 2009, 1789, 518–528. [Google Scholar] [CrossRef]
- Lin, J.Y.; Li, M.L.; Huang, P.N.; Chien, K.Y.; Horng, J.T.; Shih, S.R. Heterogeneous nuclear ribonuclear protein k interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J. Gen. Virol. 2008, 89, 2540–2549. [Google Scholar] [CrossRef]
- Huang, H.I.; Chang, Y.Y.; Lin, J.Y.; Kuo, R.L.; Liu, H.P.; Shih, S.R.; Wu, C.C. Interactome analysis of the ev71 5′ untranslated region in differentiated neuronal cells sh-sy5y and regulatory role of fbp3 in viral replication. Proteomics 2016, 16, 2351–2362. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-T.; Kung, Y.-A.; Li, M.-L.; Brewer, G.; Lee, K.-M.; Liu, S.-T.; Shih, S.-R. Additive promotion of viral internal ribosome entry site-mediated translation by far upstream element-binding protein 1 and an enterovirus 71-induced cleavage product. PLoS Pathog. 2016, 12, e1005959. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Shih, S.-R.; Pan, M.; Li, C.; Lue, C.-F.; Stollar, V.; Li, M.-L. Hnrnp a1 interacts with the 5′ untranslated regions of enterovirus 71 and sindbis virus rna and is required for viral replication. J. Virol. 2009, 83, 6106–6114. [Google Scholar] [CrossRef]
- Noah, J.W.; Shapkina, T.; Wollenzien, P. Uv-induced crosslinks in the 16s rrnas of escherichia coli, bacillus subtilis and thermus aquaticus and their implications for ribosome structure and photochemistry. Nucleic Acids Res. 2000, 28, 3785–3792. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.; Fry, E.; Stuart, D.; Lyons, C.; Hoey, E.; Martin, S.J. Preliminary crystallographic analysis of bovine enterovirus. J. Mol. Biol. 1993, 231, 930–932. [Google Scholar] [CrossRef] [PubMed]
- Poyry, T.; Kinnunen, L.; Hyypia, T.; Brown, B.; Horsnell, C.; Hovi, T.; Stanway, G. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 1996, 77, 1699–1717. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.; Tate, J.; Hoey, E.; Lyons, C.; Martin, S.; Stuart, D. Implications for viral uncoating from the structure of bovine enterovirus. Nat. Struct. Biol. 1995, 2, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Racaniello, V.R. Enterovirus 70 receptor utilization is controlled by capsid residues that also regulate host range and cytopathogenicity. J. Virol. 2007, 81, 8648–8655. [Google Scholar] [CrossRef] [PubMed]
- Olson, N.H.; Kolatkar, P.R.; Oliveira, M.A.; Cheng, R.H.; Greve, J.M.; McClelland, A.; Baker, T.S.; Rossmann, M.G. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA 1993, 90, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Karnauchow, T.M.; Tolson, D.L.; Harrison, B.A.; Altman, E.; Lublin, D.M.; Dimock, K. The hela cell receptor for enterovirus 70 is decay-accelerating factor (cd55). J. Virol. 1996, 70, 5143–5152. [Google Scholar] [PubMed]
- Smyth, M.S.; Trudgett, A.; Hoey, E.M.; Martin, S.J.; Brown, F. Characterization of neutralizing antibodies to bovine enterovirus elicited by synthetic peptides. Arch. Virol. 1992, 126, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.J.; Kremer, M.J.; Luo, M.; Vriend, G.; Arnold, E.; Kamer, G.; Rossmann, M.G.; McKinlay, M.A.; Diana, G.D.; Otto, M.J. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 1986, 233, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Arnold, E.; Rossmann, M.G. The use of molecular-replacement phases for the refinement of the human rhinovirus 14 structure. Acta Cryst. 1988, 44, 270–283. [Google Scholar] [CrossRef]
- Kim, S.S.; Smith, T.J.; Chapman, M.S.; Rossmann, M.C.; Pevear, D.C.; Dutko, F.J.; Felock, P.J.; Diana, G.D.; McKinlay, M.A. Crystal structure of human rhinovirus serotype 1a (hrv1a). J. Mol. Biol. 1989, 210, 91–111. [Google Scholar] [CrossRef]
- Sun, D.; Chen, S.; Cheng, A.; Wang, M. Roles of the picornaviral 3c proteinase in the viral life cycle and host cells. Viruses 2016, 8, 82. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Wang, J.; Fan, T.; Qin, B.; Guo, L.; Lei, X.; Wang, J.; Wang, M.; Jin, Q. Crystal structure of human enterovirus 71 3c protease. J. Mol. Biol. 2011, 408, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.; Pathak, H.B.; Cameron, C.E.; Rombaut, B.; Wimmer, E.; Paul, A.V. Stimulation of poliovirus synthesis in a hela cell-free in vitro translation-rna replication system by viral protein 3cdpro. J. Virol. 2005, 79, 6358–6367. [Google Scholar] [CrossRef]
- Blair, W.S.; Parsley, T.B.; Bogerd, H.P.; Towner, J.S.; Semler, B.L.; Cullen, B.R. Utilization of a mammalian cell-based rna binding assay to characterize the rna binding properties of picornavirus 3c proteinases. RNA 1998, 4, 215–225. [Google Scholar]
- Pecson, B.M.; Martin, L.V.; Kohn, T. Quantitative pcr for determining the infectivity of bacteriophage ms2 upon inactivation by heat, uv-b radiation, and singlet oxygen: Advantages and limitations of an enzymatic treatment to reduce false-positive results. Appl. Environ. Microbiol. 2009, 75, 5544–5554. [Google Scholar] [CrossRef]
- Templeton, M.R.; Andrews, R.C.; Hofmann, R. Inactivation of particle-associated viral surrogates by ultraviolet light. Water Res. 2005, 39, 3487–3500. [Google Scholar] [CrossRef]
- Barrett, M.; Fitzhenry, K.; O'Flaherty, V.; Dore, W.; Keaveney, S.; Cormican, M.; Rowan, N.; Clifford, E. Detection, fate and inactivation of pathogenic norovirus employing settlement and uv treatment in wastewater treatment facilities. Sci. Total Environ. 2016, 568, 1026–1036. [Google Scholar] [CrossRef]
- Klöcking, R.; Felber, Y.; Guhr, M.; Meyer, G.; Schubert, R.; Schoenherr, J.I. Development of an innovative peat lipstick based on the uv-b protective effect of humic substances. Mires Peat 2013, 11, 1–9. [Google Scholar]
- Westerhoff, P.; Aiken, G.; Amy, G.; Debroux, J. Relationships between the structure of natural organic matter and its reactivity towards molecular ozone and hydroxyl radicals. Water Res. 1999, 33, 2265–2276. [Google Scholar] [CrossRef]
- Sun, C.-X.; Kitajima, M.; Gin, K.Y.-H. Sunlight inactivation of somatic coliphage in the presence of natural organic matter. Sci. Total Environ. 2016, 541, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Symonds, E.M.; Griffin, D.W.; Breitbart, M. Eukaryotic viruses in wastewater samples from the united states. Appl. Environ. Microbiol. 2009, 75, 1402–1409. [Google Scholar] [CrossRef]
- Bofill-Mas, S.; Albinana-Gimenez, N.; Clemente-Casares, P.; Hundesa, A.; Rodriguez-Manzano, J.; Allard, A.; Calvo, M.; Girones, R. Quantification and stability of human adenoviruses and polyomavirus jcpyv in wastewater matrices. Appl. Environ. Microbiol. 2006, 72, 7894–7896. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, K.G.; Conroy, R.M.; Mosler, H.-J.; Preez, M.d.; Ubomba-Jaswa, E.; Fernandez-Ibañez, P. Solar water disinfection (sodis): A review from bench-top to roof-top. J. Hazard. Mater. 2012, 235–236, 29–46. [Google Scholar] [CrossRef]
- Canonica, S.; Kohn, T.; Mac, M.; Real, F.J.; Wirz, J.; von Gunten, U. Photosensitizer method to determine rate constants for the reaction of carbonate radical with organic compounds. Environ. Sci. Technol. 2005, 39, 9182–9188. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).