Bio-Methanol Production Using Treated Domestic Wastewater with Mixed Methanotroph Species and Anaerobic Digester Biogas
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation of Methanotrophs
2.2. Treated Wastewater in the Wastewater Treatment Plant
2.3. Biogas in the Wastewater Treatment Plant
2.4. Biogas and Metabolite Measurements
2.5. Enzyme Assays
2.6. Microbial Community Analysis
2.7. Water Quality Analysis
2.8. Statistical Analysis
3. Results and Discussion
3.1. Analysis of Cultured Methanotrophs
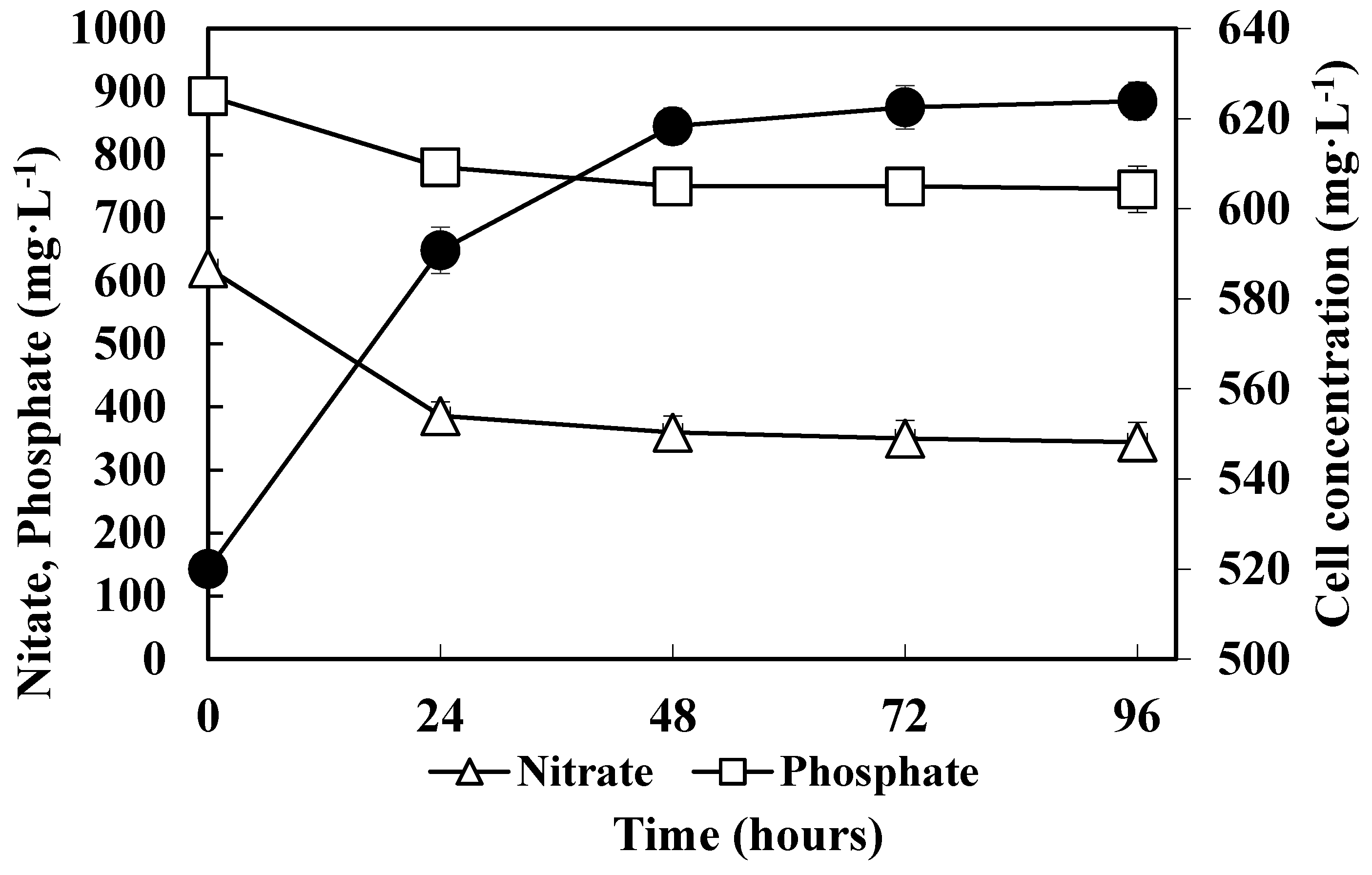
3.2. Consumption of Nitrate and Phosphate by Methanotrophs
3.3. Comparison of the Methanotroph Growth Rate and Methane Absorption Rate Using Different Culture Solutions
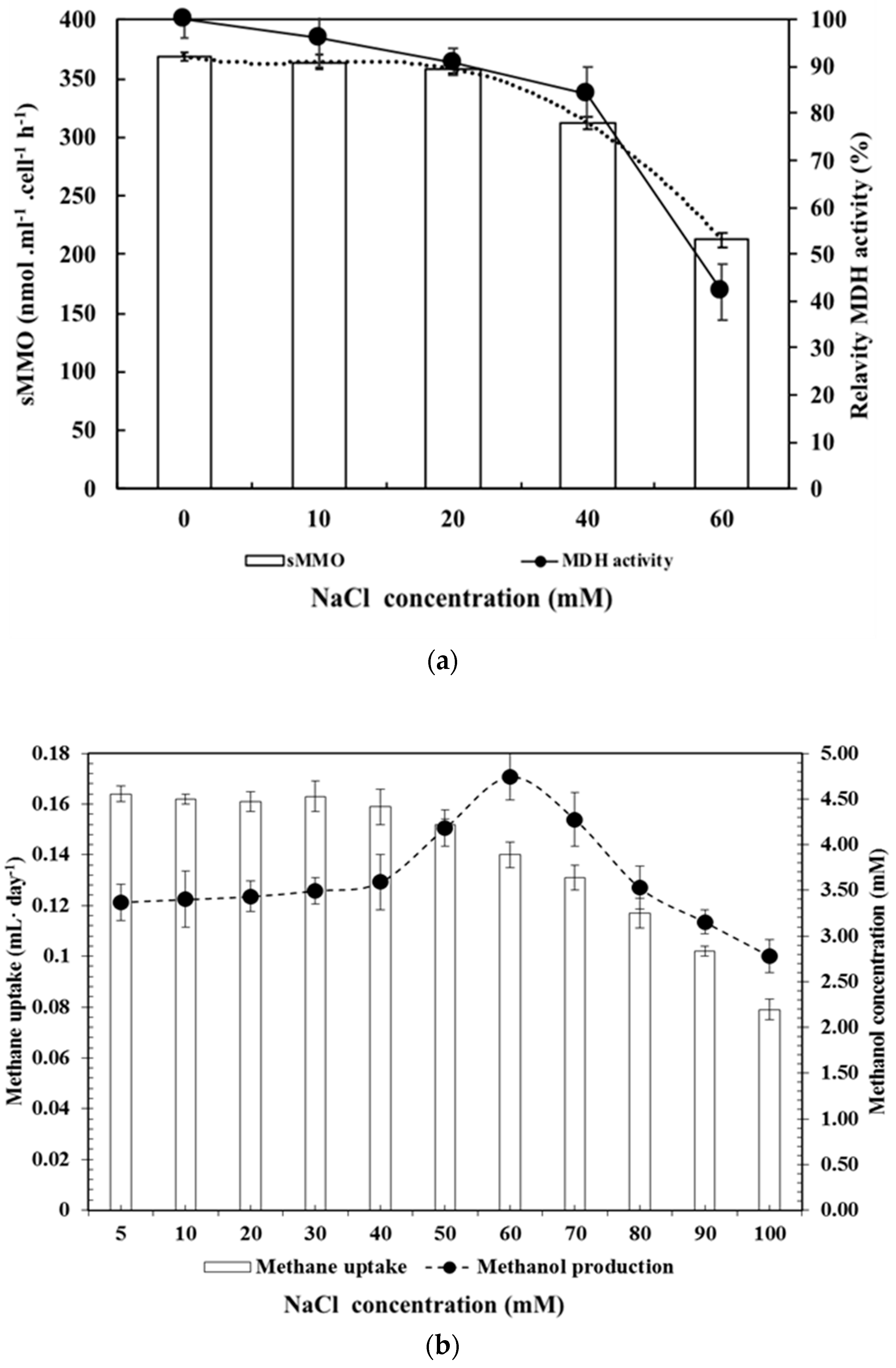
3.4. Effect of NaCl as an MDH Inhibitor, Including Analysis of Methane Uptake and Methanol Production
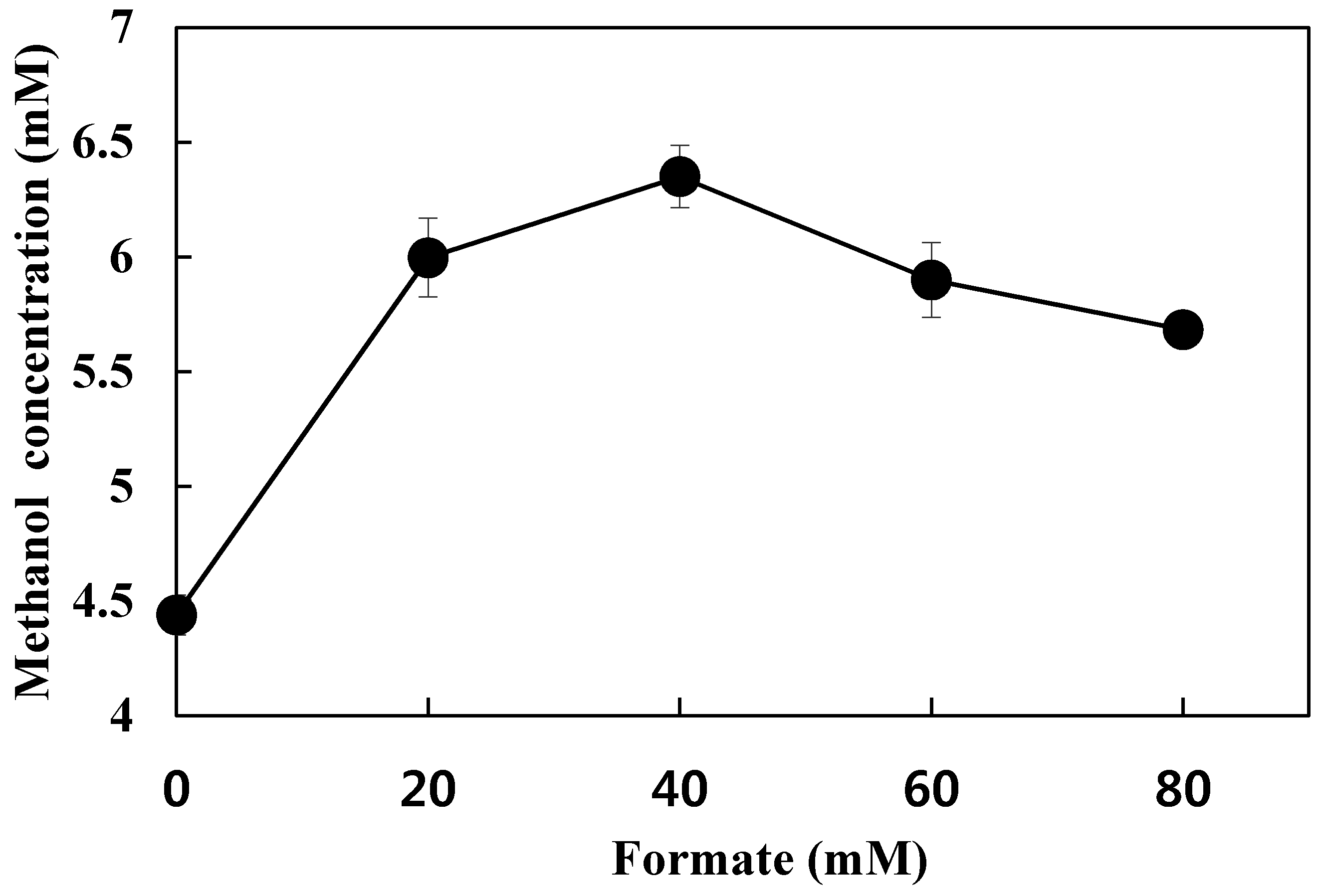
3.5. Effect of Formate
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- NCSL (National Conference of State Legislatures). Overview of the Water-Energy Nexus in the United States. 2014. Available online: http://goo.gl/2pFxgQ (accessed on 3 October 2018).
- Stillwell, A.S.; Hoppock, D.C.; Webber, M.E. Energy Recovery from Wastewater Treatment Plants in the United States: A Case Study of the Energy-Water Nexus. Sustainability 2010, 2, 945–962. [Google Scholar] [CrossRef]
- Choi, Y.J. Urban Waste Water Treatment, Move beyond Sewage Treatment to Energy Production Bases; Water Regeneration Center, Seoul’s Top Policy Package: Seoul, Korea, 2015; pp. 1–26. [Google Scholar]
- Basic Energy Independence Plan for Sewage Treatment Plant; Ministry of Environment: Seoul, Korea, 2010.
- Chan, H.T.C.; Anthony, C. The mechanism of inhibition by EDTA and EGTA of methanol oxidation by methyltrophic bacteria. FEMS Microbiol. Lett. 1992, 96, 231–234. [Google Scholar] [CrossRef]
- Duan, C.; Luo, M.; Xing, X. High-rate conversion of methane to methanol by Methylosinus trichosporium OB3b. Bioresour. Technol. 2011, 102, 7349–7353. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Ahn, C.M.; Mahanty, B.; Kim, C.G. Partial oxidative conversion of methane to methanol through selective inhibition of methanol dehydrogenase in methanotrophic consortium from landfill cover soil. Appl. Biochem. Biotechnol. 2013, 171, 1487–1499. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.Y.; Lee, S.H.; Choi, Y.S.; Park, S.J.; Na, J.G.; Chang, I.S.; Kim, C.K.; Kim, H.C.; Kim, Y.H.; Lee, J.W.; et al. Biocatalytic conversion of methane to methanol as a key step for development of methane-based biorefineries. J. Microbiol. Biotechnol. 2014, 24, 1597–1605. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.G.; Han, G.H.; Kim, S.W. Optimization of lab scale methanol production by Methylosinus trichosporium OB3b. Biotechnol. Bioprocess Eng. 2010, 15, 476–480. [Google Scholar] [CrossRef]
- Mardina, P.; Li, J.; Patel, S.K.S.; Kim, I.W.; Lee, J.K.; Selvara, C. Potential of immobilized whole-cell Methylocella tundrae as a biocatalyst for methanol production from methane. J. Microbiol. Biotechnol. 2016, 26, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.K.; Mardina, P.; Kim, S.Y.; Lee, J.K.; Kim, I.W. Biological methanol production by a Type II methanotroph Methylocystis bryophila. J. Microbiol. Biotechnol. 2016, 26, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.S.; Han, J.S.; Ahn, C.M.; Min, D.H.; Mo, W.J.; Yoon, S.U.; Lee, J.G.; Lee, J.Y.; Kim, C.G. Characteristics of methanol production derived from methane oxidation by inhibiting methanol dehydrogenase. J. Korea Soc. Environ. Eng. 2011, 33, 662–669. [Google Scholar] [CrossRef]
- Higgins, I.J.; Best, D.J.; Hammond, R.C.; Scott, D. Methane-oxidizing microorganisms. Microbiol. Rev. 1981, 45, 556–590. [Google Scholar] [PubMed]
- Sheets, J.P.; Ge, X.; Li, Y.F.; Yu, Z.; Li, Y. Biological conversion of biogas to methanol using methanotrophs isolated from solid-state anaerobic digestate. Bioresour. Technol. 2016, 201, 50–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patal, S.K.S.; Jeong, J.H.; Mehariya, S.; Sachin, V.; Otari, S.V.; Madan, B.; Hwa, J.R. Production of Methanol from Methane by Encapsulated Methylosinus sporium. J. Microbiol. Biotechnol. 2016, 26, 2098–2105. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.K.S.; Kumar, V.; Mardina, P.; Li, J.; Lestari, R.; Kalia, V.C.; Lee, J.K. Methanol production from simulated biogas mixtures by co-immobilized Methylomonas methanica and Methylocella tundrae. Bioresour Technol. 2018, 263, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Hoefman, S.; Ha, D.; van der Boon, N.; Vandamme, P.; Vos, P.D.; Kim, H. Niche differentiation in nitrogen metabolism among methanotrophs within an operational taxonomic unit. BMC Microbiol. 2014, 14, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedard, C.; Knowles, R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 1989, 53, 68–84. [Google Scholar] [PubMed]
- Calsen, H.N.; Joergensen, L.; Degn, H. Inhibition by ammonia of methane utilization in Methylococcus capsulatus (Bath). Appl. Microbiol. Biotechnol. 1991, 35, 124–127. [Google Scholar]
- Bradford, M.A.; Ineson, P.; Wookey, P.A.; Lappin-Scott, H.M. The effects of acid nitrogen and acid sulphur deposition on CH4 oxidation in a forest soil: A laboratory study. Soil Biol. Biochem. 2001, 33, 1695–1702. [Google Scholar] [CrossRef]
- Wang, Z.P.; Ineson, P. Methane oxidation in a temperate coniferous forest soil: Effects of inorganic N. Soil Biol. Biochem. 2003, 35, 427–433. [Google Scholar] [CrossRef]
- Schnell, S.; King, G.M. Mechanistic analysis of ammonium inhibition of atmospheric methane consumption in forest soil. Appl. Environ. Microbiol. 1994, 60, 3514–3521. [Google Scholar] [PubMed]
- Semrau, J.D.; DiSpirito, A.A.; Yoon, S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010, 34, 496–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brusseau, G.A.; Tsien, H.C.; Hanson, R.S.; Wackett, L.P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation 1990, 1, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Hristova, K.R.; Lidstrom, M.E.; Chistoserdova, L. Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: Implications for environmental detection of methylotrophy and evidence for convergent evolution. J. Bacteriol. 2008, 190, 3817–3823. [Google Scholar] [CrossRef] [PubMed]
- Madigan, M.T.; Martino, J.M. Brock Biology of Microorganisms, 11th ed.; Pearson: London, UK, 2006; p. 136. [Google Scholar]
- Takeuchi, M.; Kamagata, Y.; Oshima, K.; Hanada, S.; Tamaki, H.; Marumo, K.; Maeda, H.; Nedachi, M.; Hattori, M.; Iwasaki, W.; et al. Methylocaldum marinum sp. nov., a novel thermotolerant methane oxidizing bacterium isolated from marine sediments. Int. J. Syst. Evol. Microbiol. 2014, 64, 3240–3246. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.; Sly, L.; Cox, J.; Nichols, P.; Hayward, A.C. Revised taxonomy of the methanotrophs: Description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Bacteriol. 1993, 43, 735–753. [Google Scholar] [CrossRef]
- Oshkin, I.; Beck, D.A.C.B.; Lamb, A.E.; Tchesnokova, V.; Benuska, G.; McTaggart, T.L.; Kalyuzhnaya, M.G.; Dedysh, S.; Lidstrom, M.E.; Chistoserdova, L. Methane fed microcosms show differential community dynamics and pinpoint specific taxa involved in communal response. ISME J. 2014, 9, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Whittenbury, R.; Phillips, K.C.; Wilkinson, J.F. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 1970, 61, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Koh, S.C.; Bowman, J.P.; Sayler, G.S. Soluble methane monooxygenase production and trichloroethylene degradation by a type I methanotroph, Methylomonas methanica 68-1. Appl. Environ. Microbiol. 1993, 59, 960–967. [Google Scholar] [PubMed]
- Collins, M.L.; Buchholz, L.A.; Remsen, C.C. Effect of copper on Methylomonas albus BG8. Appl. Environ. Microbiol. 1991, 57, 1261–1264. [Google Scholar] [PubMed]
- Lemos, S.S.; Perille Collins, M.L.; Eaton, S.S.; Eaton, G.R.; Antholine, W.E. Comparison of EPR-visible Cu (2+) sites in pMMO from Methylococcus capsulatus (Bath) and Methylomicrobium album BG8. Biophys. J. 2000, 79, 1085–1094. [Google Scholar] [CrossRef]
- Lidstrom, M.E. Isolation and characterization of marine methanotrophs. Antonie Van Leeuwenhoek 1988, 54, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, Y.Z. Khalifa, Mutagenesis of a Copper P-Type ATPase Encoding Gene in Methylococcus capsulatus (Bath) Results in Copper-Resistance. Int. J. Biosci. Biochem. Bioinform. 2013, 3, 37–42. [Google Scholar]
- Khalifa, A.Y.Z. Physiological features during the copper switch in Methylococcus capsulatus (Bath). Int. J. Biosci. Biochem. Bioinform. 2013, 3, 63–66. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martnko, J.M.; Parker, J. Biology of Microorganisms, 8th ed.; Simon & Schuster: New York, NY, USA, 1997. [Google Scholar]
- Avdeeva, L.; Gvozdev, R. Effect of copper concentration on the growth of Methylococcus capsulatus (Strain M). Chem. J. Mold. 2017, 12, 110–114. [Google Scholar] [CrossRef]
- Anthony, C. The Biochemistry of Methylotrophs; Academic Press Ltd.: London, UK, 1982. [Google Scholar]
- Xin, J.Y.; Cui, J.R.; Niu, J.Z.; Hua, S.F.; Xia, C.G.; Li, S.B.; Zhu, L.M. Production of methanol from methane by methanotrophic bacteria. Biocatal. Biotransform. 2004, 22, 225–229. [Google Scholar] [CrossRef]
- Yoo, Y.S.; Han, J.S.; Ahn, C.M.; Kim, C.G. Comparative enzyme inhibitive methanol production by Methylosinus sporium from simulated biogas. J. Environ. Technol. 2015, 36, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.G.; Goo, J.H.; Kim, H.G.; Oh, J.L.; Kim, Y.M.; Kim, S.W. Optimization of methanol biosynthesis from methane using Methylosinus trichosporium OB3b. Biotechnol. Lett. 2004, 26, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.G.; Kim, S.W. Purification and characterization of a methanol dehydrogenase derived from Methylomicrobium sp. HG-1 cultivated using a compulsory circulation diffusion system. Biotechnol. Bioprocess Eng. 2006, 11, 134–139. [Google Scholar] [CrossRef]
- Palumbo, A.V.; Strong-Gunderson, M.; Carroll, S. Retaining and recovering enzyme activity during degradation of TCE by methanotrophs. Appl. Biochem. Biotechnol. 1997, 63–65, 789–796. [Google Scholar] [CrossRef] [PubMed]
- McDonald, I.R.; Miguez, C.B.; Rogge, G.; Bourque, D.; Wendlandt, K.D.; Groleau, D.; Murrell, J.C. Diversity of soluble methane monooxygenase-containing methanotrophs isolated from polluted environments. Microbiol. Lett. 2006, 255, 225–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Species | MMO Type | ICM Type | Reference |
|---|---|---|---|
| Methylomonas methanica | pMMO, sMMO | Type I | [26] |
| Methylococcus capsulatus | pMMO, sMMO | Type X | [22] |
| Methylococcus bovis | pMMO | Type I | [23] |
| Methylobacter marinus | pMMO, sMMO | Type I | [27] |
| (1) Metabolite Production | ||||
| MDH Control Condition (mM) | Methanol | Formaldehyde | Formate | Total Metabolites |
| Control | 3.46 (±0.17) | 2.04 (±0.13) | 0.80 (±0.09) | 6.30 (±0.18) |
| NaCl 60 mM | 4.52 (±0.19) | 0.81 (±0.11) | 0.13 (±0.03) | 5.46 (±0.13) |
| NaCl 60 mM + sodium formate 40 mM | 6.35 (±0.14) | 0.72 (±0.12) | - | - |
| (2) Comparison of Methanol Production in Various Studies Using NaCl as an MDH Inhibitor | ||||
| MDH Control Condition | Methanol (mM) | Methanotroph Species | Reference | |
| 50 mM MgCl2 50 mM NaCl + 10 mM NH4Cl 50 mM MgCl2 + 100 mM sodium formate | 2.21 mM <2.2 mM 4.15 mM | Methylocystis bryophila | [11] | |
| 1 mM EDTA + 100 mM NaCl + 20 mM sodium formate | 13.2 mM | Methylosinus trihcosporium OB3b | [9] | |
| NaCl (100 mM) 40 mM (NH4Cl) | 0.60 mmol (6.0 mM) 0.74 mmol (7.4 mM) | Methylosinus sporium | [41] | |
| 200 mM NaCl + 20 mM sodium formate | 7.7 mM | Methylosinus trichosporium OB3b | [42] | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, I.-T.; Yoo, Y.-S.; Yoon, Y.-H.; Lee, Y.-E.; Jo, J.-H.; Jeong, W.; Kim, K.-S. Bio-Methanol Production Using Treated Domestic Wastewater with Mixed Methanotroph Species and Anaerobic Digester Biogas. Water 2018, 10, 1414. https://doi.org/10.3390/w10101414
Kim I-T, Yoo Y-S, Yoon Y-H, Lee Y-E, Jo J-H, Jeong W, Kim K-S. Bio-Methanol Production Using Treated Domestic Wastewater with Mixed Methanotroph Species and Anaerobic Digester Biogas. Water. 2018; 10(10):1414. https://doi.org/10.3390/w10101414
Chicago/Turabian StyleKim, I-Tae, Young-Seok Yoo, Young-Han Yoon, Ye-Eun Lee, Jun-Ho Jo, Wonsik Jeong, and Kwang-Soo Kim. 2018. "Bio-Methanol Production Using Treated Domestic Wastewater with Mixed Methanotroph Species and Anaerobic Digester Biogas" Water 10, no. 10: 1414. https://doi.org/10.3390/w10101414
APA StyleKim, I.-T., Yoo, Y.-S., Yoon, Y.-H., Lee, Y.-E., Jo, J.-H., Jeong, W., & Kim, K.-S. (2018). Bio-Methanol Production Using Treated Domestic Wastewater with Mixed Methanotroph Species and Anaerobic Digester Biogas. Water, 10(10), 1414. https://doi.org/10.3390/w10101414





