Synteny-Based Development of CAPS Markers Linked to the Sweet kernel LOCUS, Controlling Amygdalin Accumulation in Almond (Prunus dulcis (Mill.) D.A.Webb)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Isolation
2.2. Simple Sequence Repeat Marker Analysis
2.3. Development of Sk-Linked CAPS Markers
2.4. Linkage Analysis
2.5. CAPS Assay on a Cultivar Collection
2.6. Bioinformatic Characterization of the Peach Sk Synthenic Region
3. Results
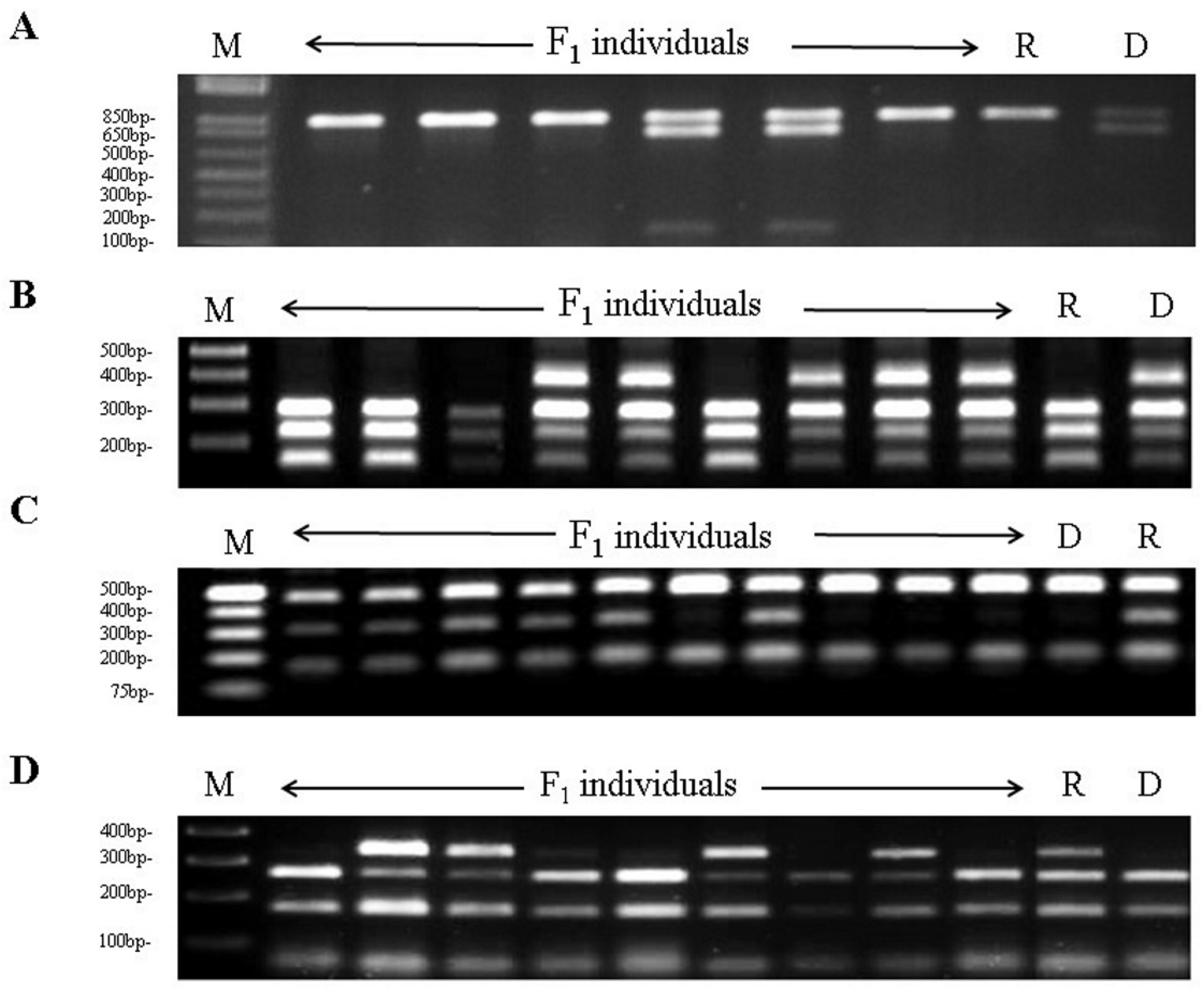
3.1. Development of Sk-Linked CAPS Markers
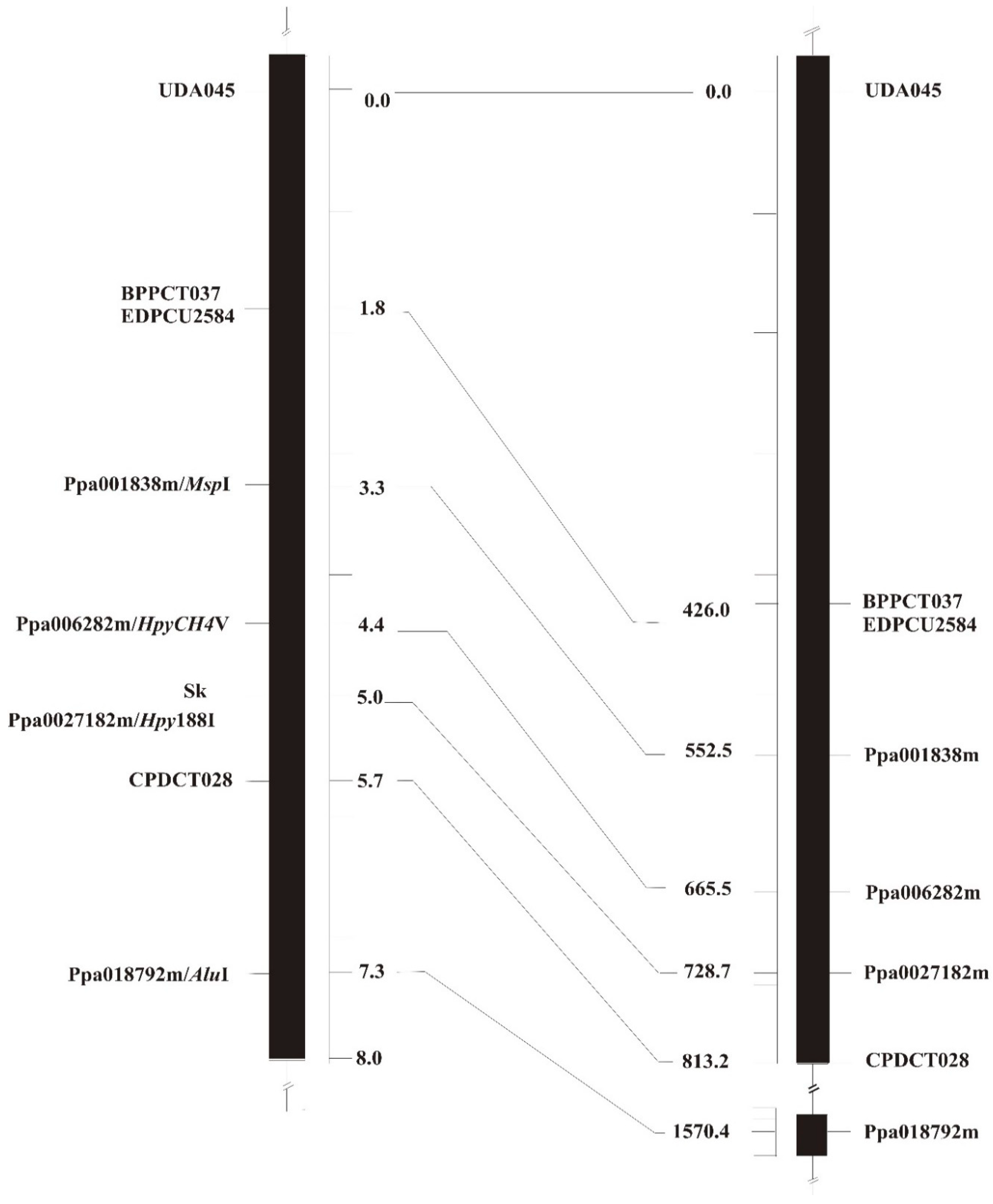
3.2. Sk Mapping and Syntenic Relationships with the Peach Genome
3.3. Marker Validation in An Almond Germplasm Collection
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Zagrobelny, M.; Bak, S.; Rasmussen, A.V.; Jørgensen, B.; Naumann, C.M.; Møller, B.L. Cyanogenic glucosides and plant-insect interactions. Phytochemistry 2004, 65, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Dicenta, F.; Ortega, E.; Martìnez-Gòmez, P. Use of recessive homozygous genotypes to assess the genetic control of kernel bitterness in almond. Euphytica 2007, 153, 221–225. [Google Scholar] [CrossRef]
- Dicenta, F.; Martìnez-Gòmez, P.; Ortega, E.; Duval, H. Cultivar pollinizer does not affect almond flavour. HortScience 2000, 35, 1153–1154. [Google Scholar]
- Sánchez-Pérez, R.; Howad, W.; Dicenta, F.; Arùs, P.; Martìnez-Gòmez, P. Mapping major genes and quantitative trait loci controlling agronomic traits in almond. Plant Breed. 2007, 126, 310–318. [Google Scholar] [CrossRef]
- Dicenta, F.; García, J.E. Inheritance of the kernel flavour in almond. Heredity 1993, 70, 308–312. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Pérez, R.; Jørgensen, K.; Olsen, C.E.; Dicenta, F.; Møller, B.L. Bitternes in almond. Plant Physiol. 2008, 146, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Pérez, R.; Belmonte, F.S.; Borch, J.; Dicenta, F.; Møller, B.L.; Jørgensen, K. Prunasin hydrolases during fruit development in sweet and bitter almonds. Plant Physiol. 2012, 158, 1916–1932. [Google Scholar] [CrossRef] [PubMed]
- Carneiro Vieira, M.L.; Santini, L.; Diniz, A.L.; de Freitas Munhoz, C. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Schiavulli, A.; Lotti, C.; Ricciardi, L. CAPS technology as a tool for the development of genic and functional markers: Study in peas. In Cleaved Amplified Polymorphic Sequences (CAPS) Markers in Plant Biology; Shavrukov, Y., Ed.; Nova Publisher: New York, NY, USA, 2014; pp. 83–90. [Google Scholar]
- Pavan, S.; Schiavulli, A.; Appiano, M.; Miacola, C.; Visser, R.G.F.; Bai, Y.; Lotti, C.; Ricciardi, L. Identification of a complete set of functional markers for the selection of er1 powdery mildew resistance in pea. Mol. Breed. 2013, 31, 247–253. [Google Scholar] [CrossRef]
- Pavan, S.; Zheng, Z.; van den Berg, P.; Lotti, C.; De Giovanni, C.; Borisova, M.; Lindhout, P.; de Jong, H.; Ricciardi, L.; Visser, R.G.F.; et al. Map vs. homology-based cloning for the recessive gene ol-2 conferring resistance to tomato powdery mildew. Euphytica 2008, 162, 91–98. [Google Scholar] [CrossRef]
- Sánchez-Pérez, R.; Howad, W.; Garcia-Mas, J.; Arús, P.; Martínez-Gómez, P.; Dicenta, F. Molecular markers for kernel bitterness in almond. Tree Genet. Genomes 2010, 6, 237–245. [Google Scholar] [CrossRef]
- Dirlewanger, E.; Cosson, P.; Howad, W.; Capdeville, G.; Bosselut, N.; Claverie, M.; Voisin, R.; Poizat, C.; Lafargue, B.; Baron, O. Microsatellite genetic linkage maps of myrobalan plum and an almond-peach hybrid—Location of root-knot nematode resistance genes. Theor. Appl. Genet. 2004, 109, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Dirlewanger, E.; Crosson, A.; Tavaud, P.; Aranzana, M.J.; Poizat, C.; Zanetto, A.; Arús, P.; Laigret, L. Development of microsatellite markers in peach and their use in genetic diversity analysis in peach and sweet cherry. Theor. Appl. Genet. 2002, 105, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Mnejja, M.; García-Mas, J.; Howad, W.; Arus, P. Development and transportability across Prunus species of 42 polymorphic almond microsatellites. Mol. Ecol. Notes 2005, 5, 531–535. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Rosen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W. JoinMap 4.0: Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma B.V.: Wageningen, The Netherlands, 2006. [Google Scholar]
- Agy, S.; Poczai, P.; Cernák, I.; Gorji, A.M.; Hegedűs, G.; Taller, J. PICcalc: An online program to calculate polymorphic information content for molecular genetic studies. Biochem. Genet. 2012, 50, 670–672. [Google Scholar] [CrossRef]
- InterPro: Protein Sequence Analysis & Classification. Available online: http://www.ebi.ac.uk/interpro/ (accessed on 20 May 2017).
- Smith, S.M.; Maughan, P.J. SNP genotyping using KASPar assays. Methods Mol. Biol. 2015, 1245, 243–256. [Google Scholar] [CrossRef] [PubMed]
| Cultivar | Origin |
|---|---|
| Del Cid | Spain |
| Ramillete | Spain |
| Atocha | Spain |
| Desmayo Largueta | Spain |
| Marcona | Spain |
| Vivot | Spain |
| Peraleja | Spain |
| Antoñeta | Spain |
| Ferragnès | France |
| Lauranne | France |
| Marta | Spain |
| R-1000 | France |
| Mono | USA |
| Tioga | USA |
| Titan | USA |
| Wawona | USA |
| Nonpareil | USA |
| Tardy-Nonpareil | USA |
| Achaak | Tunisia |
| Ardechoise | France |
| Chellaston | Australia |
| Primorskii | Russia |
| Garrigues | Spain |
| Genco | Italy |
| Tuono | Italy |
| Marker | Primer Sequences (5′-3′) | PCR Product (bp) | SNP | Digestion Products (bp) |
|---|---|---|---|---|
| ppa001838m/MspI | F: GGTTGTTCTGGGAGATGGAA R: ACTTGACCGCAACCAAAATC | 800 | T→G | D: 800, 650, 150 |
| R: 800 | ||||
| ppa006282m/HpyCH4V | F: GTTTCGCTCGATTGGGTCTC R: ATCATTTCCCGCCTGAATGC | 700 | G→A | D: 400, 300, 250, 150 |
| R:300, 250, 150 | ||||
| ppa027182m/Hpy188I | F: AAAGAAGATTGGGGCCTTGT R: TGGTTAAGCTTCTCGCGTCT | 600 | C→T | D: 450, 150 |
| R: 450, 300, 150, | ||||
| ppa018792m/AluI | F: ACGTTGTCTCGTTCGTGGTT R: AGGTGCTGCAAAGACACTGA | 540 | T→C | D:280, 180, 80 |
| R:340, 280, 180, 80 |
| GDR ID | Interval on Scaffold 5 | InterPro Putative Function |
|---|---|---|
| ppa006282m | 12.547.702-12.551.295 | Uncharacterised protein family UPF0017, hydrolase-like, conserved site |
| ppa005470m | 12.555.940-12.558.349 | Cys/Met metabolism, pyridoxal phosphate-dependent enzyme |
| ppa003882m | 12.562.194-12.564.053 | Cytochrome P450 |
| ppa011942m | 12.576.856-12.578.103 | Mediator complex, subunit Med10 |
| ppa023406m | 12.587.426-12.589.276 | Glyoxal oxidase |
| ppa022201m | 12.597.330-12.598.918 | Helix-loop-helix DNA-binding-Transcription factor MYC/MYB |
| ppa025417m | 12.603.688-12.605.325 | Helix-loop-helix DNA-binding-Transcription factor MYC/MYB |
| ppa027182m | 12.612.821-12.614.522 | Helix-loop-helix DNA-binding-Transcription factor MYC/MYB |
| ppa015634m | 12.625.785-12.627.695 | Helix-loop-helix DNA-binding-Transcription factor MYC/MYB |
| ppa005343m | 12.636.946-12.638.591 | Helix-loop-helix DNA-binding-Transcription factor MYC/MYB |
| ppa005388m | 12.644.406-12.646.982 | Alpha/beta hydrolase fold-1 |
| ppa021506m | 12.649.801-12.651.659 | GDSL lipase |
| ppa010428m | 12.662.679-12.664.291 | Domain of unknown function DUF4033 |
| ppa004653m | 12.666.017-12.668.186 | Glycoside hydrolase, family 9 |
| ppa005847m | 12.669.378-12.670.953 | Transmembrane receptor, eukaryota |
| ppa022759m | 12.673.586-12.675.114 | Unknown |
| ppa019752m | 12.677.592-12.679.579 | WRC domain protein |
| ppa021141m | 12.680.973-12.682.964 | IQ motif, EF-hand binding site |
| ppa019815m | 12.687.460-12.688.443 | Glutaredoxin |
| ppa006801m | 12.695.757-12.697.169 | No apical meristem (NAM) protein |
| CPDCT028 | 12.699.037-12.699.598 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricciardi, F.; Del Cueto, J.; Bardaro, N.; Mazzeo, R.; Ricciardi, L.; Dicenta, F.; Sánchez-Pérez, R.; Pavan, S.; Lotti, C. Synteny-Based Development of CAPS Markers Linked to the Sweet kernel LOCUS, Controlling Amygdalin Accumulation in Almond (Prunus dulcis (Mill.) D.A.Webb). Genes 2018, 9, 385. https://doi.org/10.3390/genes9080385
Ricciardi F, Del Cueto J, Bardaro N, Mazzeo R, Ricciardi L, Dicenta F, Sánchez-Pérez R, Pavan S, Lotti C. Synteny-Based Development of CAPS Markers Linked to the Sweet kernel LOCUS, Controlling Amygdalin Accumulation in Almond (Prunus dulcis (Mill.) D.A.Webb). Genes. 2018; 9(8):385. https://doi.org/10.3390/genes9080385
Chicago/Turabian StyleRicciardi, Francesca, Jorge Del Cueto, Nicoletta Bardaro, Rosa Mazzeo, Luigi Ricciardi, Federico Dicenta, Raquel Sánchez-Pérez, Stefano Pavan, and Concetta Lotti. 2018. "Synteny-Based Development of CAPS Markers Linked to the Sweet kernel LOCUS, Controlling Amygdalin Accumulation in Almond (Prunus dulcis (Mill.) D.A.Webb)" Genes 9, no. 8: 385. https://doi.org/10.3390/genes9080385





