Predicting Variation of DNA Shape Preferences in Protein-DNA Interaction in Cancer Cells with a New Biophysical Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. ChIP-Seq Data
2.2. Allele-Specific Binding Data
2.3. BayesPI Transcription Factor–DNA Binding Affinity Model
2.4. DNA Shape Affinity Model
2.5. Bayesian Inference of Model Parameters for the DNA Shape Restricted Dinucleotide Dependence Model
2.6. Mutation Effect Prediction by Using Various Biophysical Models
2.7. Code Availability
3. Results
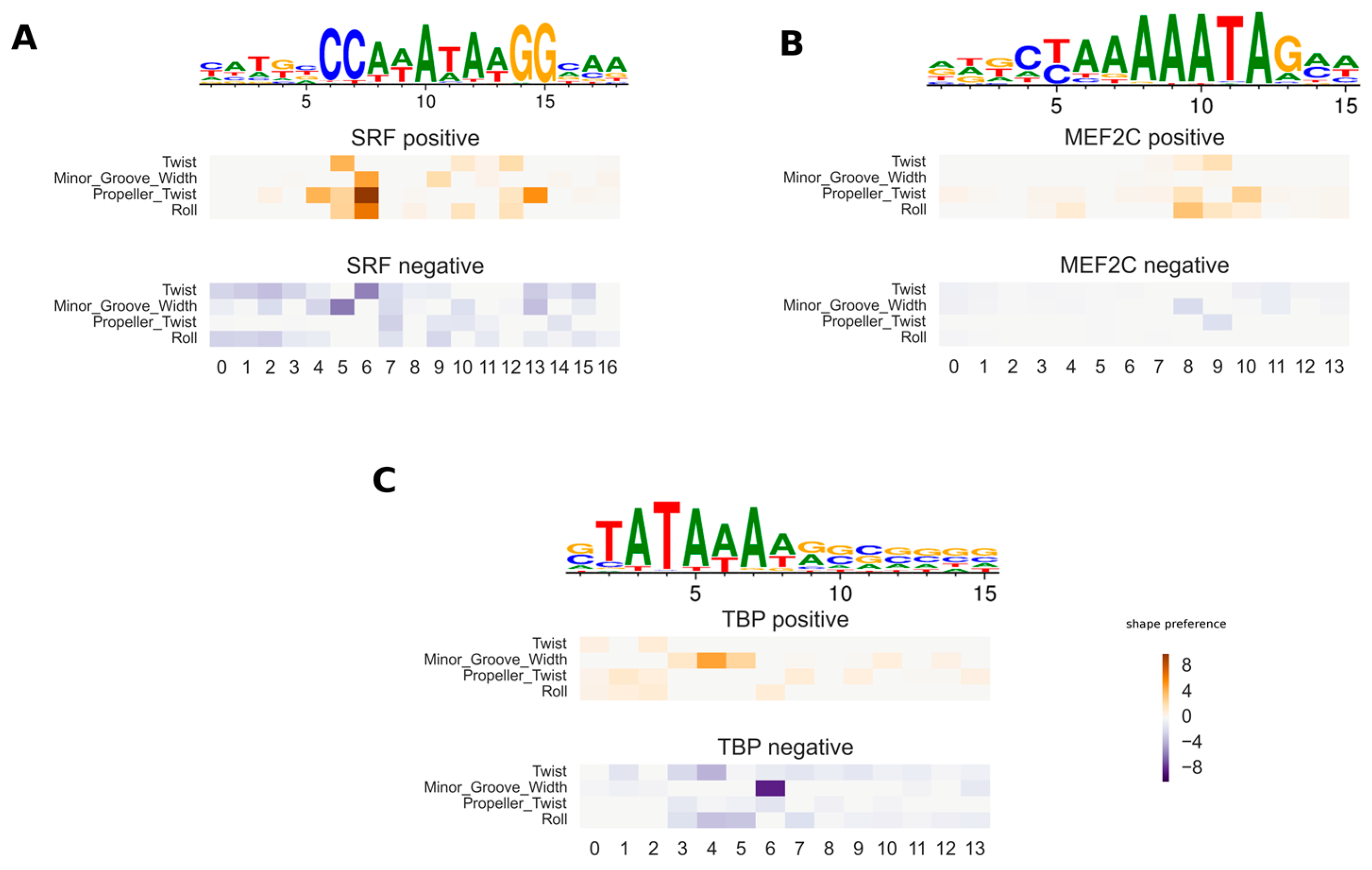
3.1. Validation of Inferred DNA Shape Feature Preferences for Protein–DNA Interaction
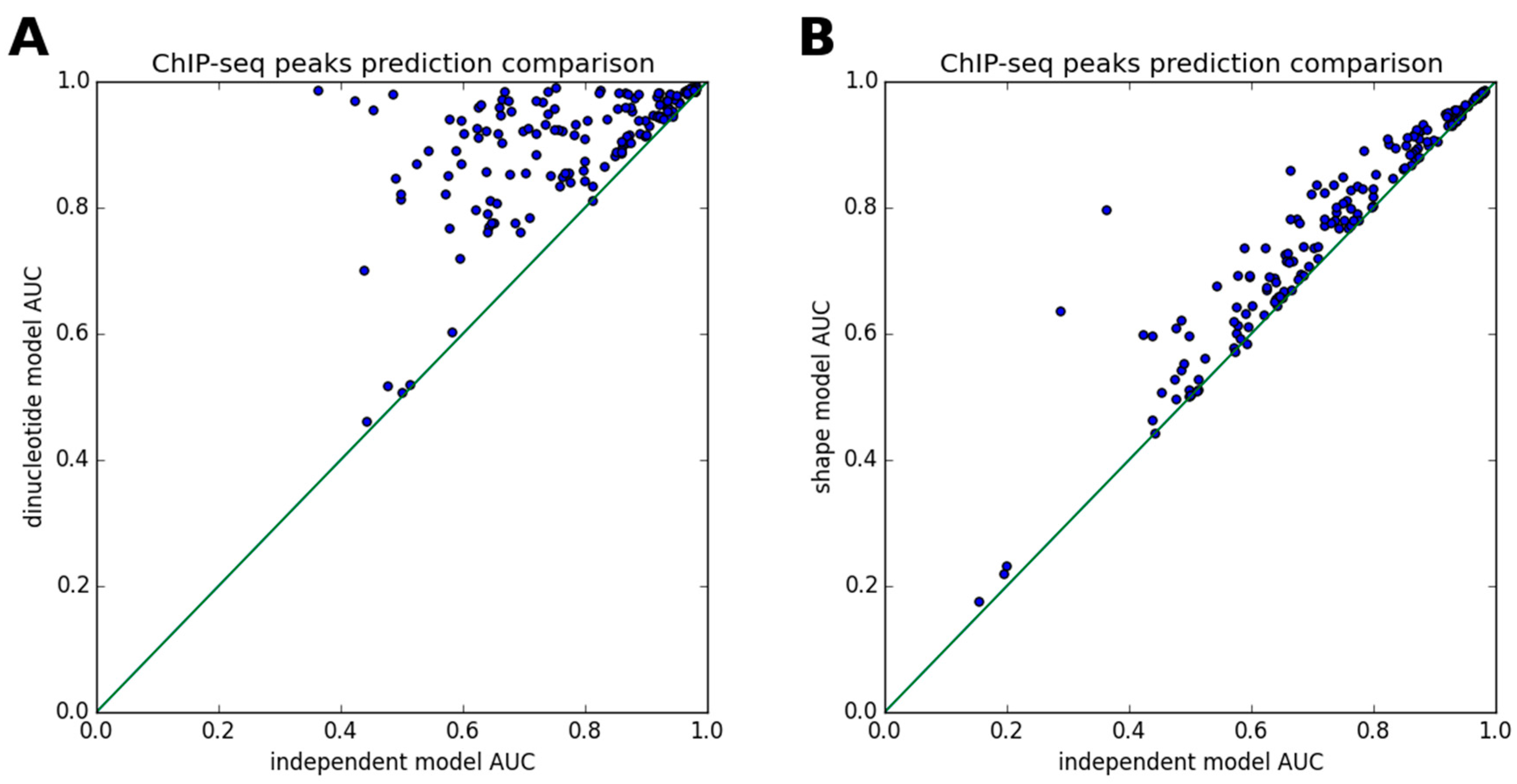
3.2. ChIP-Seq Peak Prediction
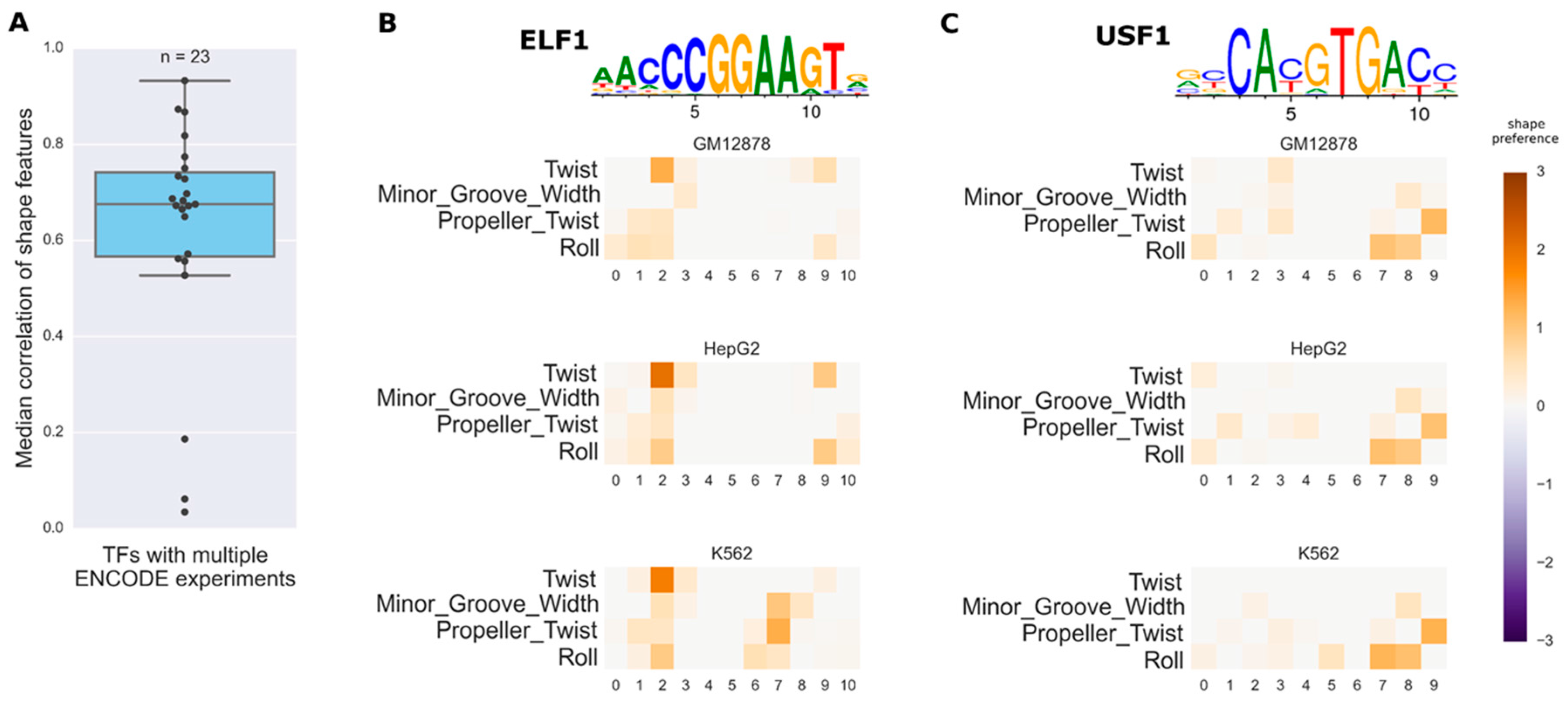
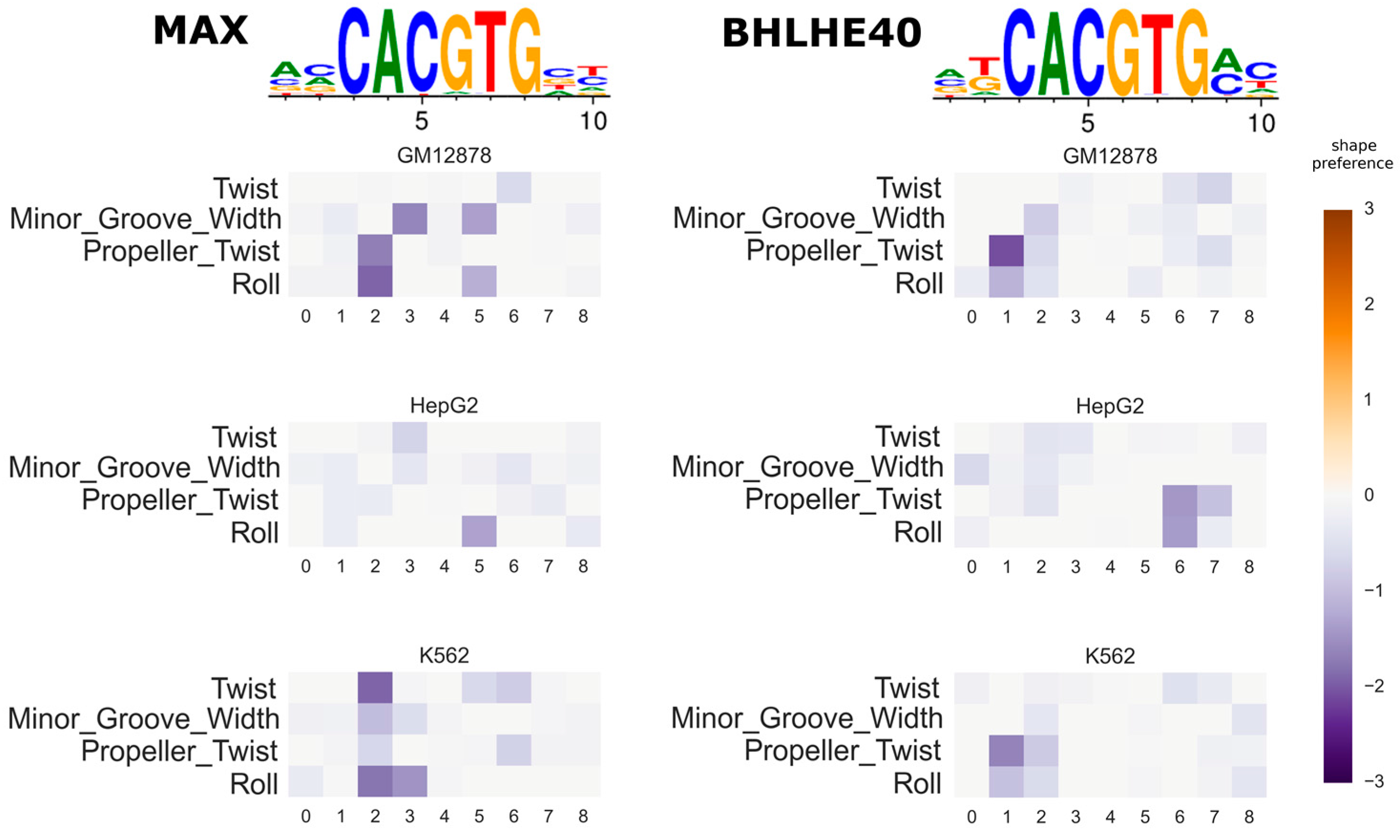
3.3. Investigating the Variation of Predicted DNA Shape Preferences across Cell Types
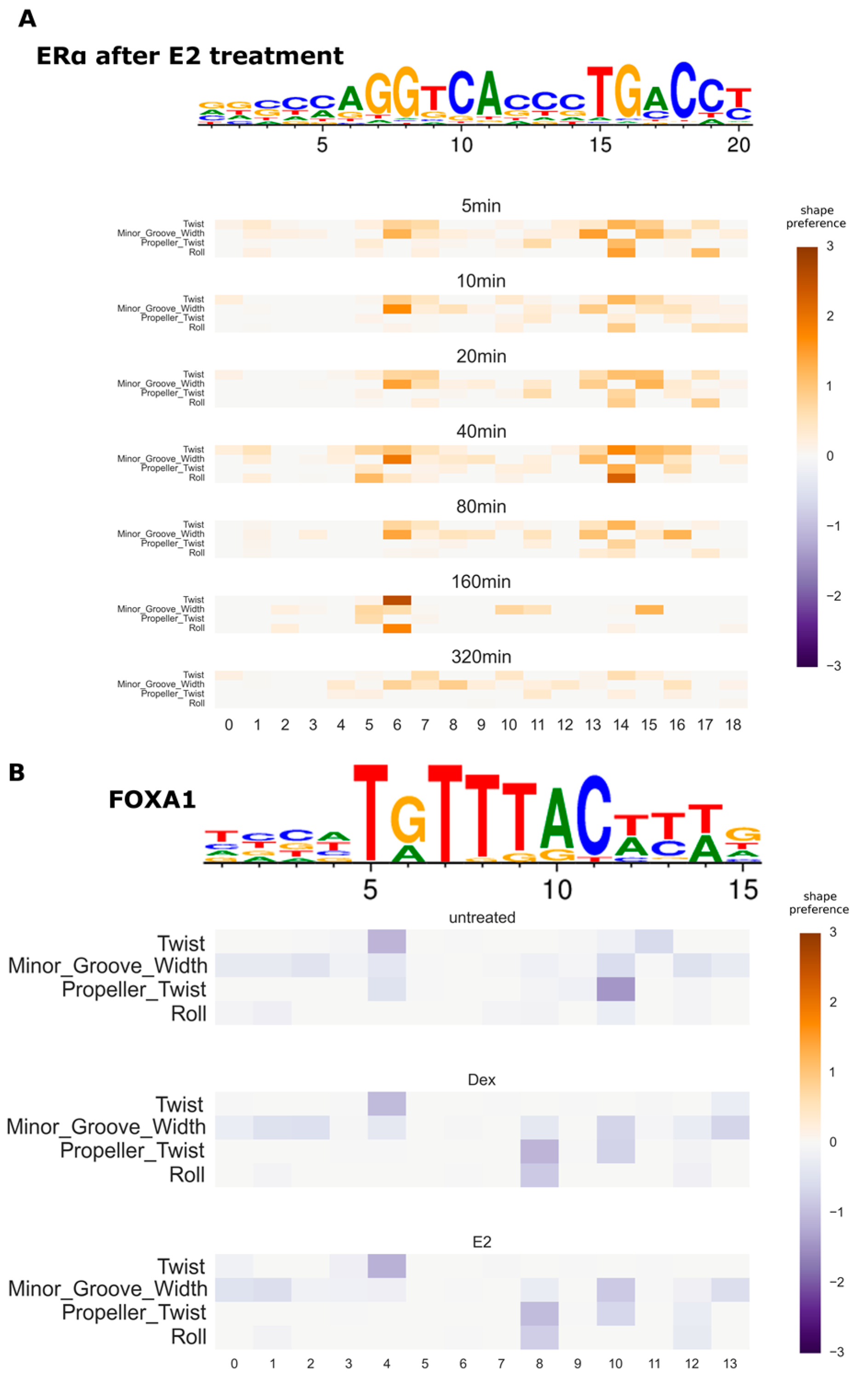
3.4. Variation of DNA Shape Preferences across Cellular Conditions
3.5. Allele-Specific Binding Prediction
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interests
References
- Slattery, M.; Zhou, T.; Yang, L.; Dantas Machado, A.C.; Gordan, R.; Rohs, R. Absence of a simple code: How transcription factors read the genome. Trends Biochem. Sci. 2014, 39, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Li, D.; Zeng, X.; Wu, Y.; Guo, L.; Zou, Q. nDNA-Prot: Identification of DNA-binding proteins based on unbalanced classification. BMC Bioinform. 2014, 15, 298. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Quality versus accuracy: Result of a reanalysis of protein-binding microarrays from the DREAM5 challenge by using BayesPI2 including dinucleotide interdependence. BMC Bioinform. 2014, 15, 289. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Batmanov, K. BayesPI-BAR: A new biophysical model for characterization of regulatory sequence variations. Nucleic Acids Res. 2015, 43, e147. [Google Scholar] [CrossRef] [PubMed]
- Abe, N.; Dror, I.; Yang, L.; Slattery, M.; Zhou, T.; Bussemaker, H.J.; Rohs, R.; Mann, R.S. Deconvolving the recognition of DNA shape from sequence. Cell 2015, 161, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Chen, W.; Qiu, C.; Wu, Y.; Krishnan, S.; Zou, Q. LibD3C: Ensemble classifiers with a clustering and dynamic selection strategy. Neurocomputing 2014, 123, 424–435. [Google Scholar] [CrossRef]
- Ghandi, M.; Lee, D.; Mohammad-Noori, M.; Beer, M.A. Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput. Biol. 2014, 10, e1003711. [Google Scholar] [CrossRef] [PubMed]
- Mathelier, A.; Wasserman, W.W. The next generation of transcription factor binding site prediction. PLoS Comput. Biol. 2013, 9, e1003214. [Google Scholar] [CrossRef] [PubMed]
- Mathelier, A.; Xin, B.; Chiu, T.P.; Yang, L.; Rohs, R.; Wasserman, W.W. DNA Shape features improve transcription factor binding site predictions in vivo. Cell Syst. 2016, 3, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Alipanahi, B.; Delong, A.; Weirauch, M.T.; Frey, B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015, 33, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Riley, T.R.; Lazarovici, A.; Mann, R.S.; Bussemaker, H.J. Building accurate sequence-to-affinity models from high-throughput in vitro protein-DNA binding data using Feature REDUCE. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ruan, S.; Pandey, M.; Stormo, G.D. Improved models for transcription factor binding site identification using nonindependent interactions. Genetics 2012, 191, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Morigen. BayesPI—A new model to study protein-DNA interactions: A case study of condition-specific protein binding parameters for Yeast transcription factors. BMC Bioinform. 2009, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Palidwor, G.A.; Perkins, T.J. BIDCHIPS: Bias decomposition and removal from ChIP-seq data clarifies true binding signal and its functional correlates. Epigenetics Chromatin 2015, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Orenstein, Y.; Shamir, R. A comparative analysis of transcription factor binding models learned from PBM, HT-SELEX and ChIP data. Nucleic Acids Res. 2014, 42, e63. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Granas, D.; Stormo, G.D. Inferring binding energies from selected binding sites. PLoS Comput. Biol. 2009, 5, e1000590. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Malecka, A.; Trøenand, G.; Delabie, J. Comprehensive genome-wide transcription factor analysis reveals that a combination of high affinity and low affinity DNA binding is needed for human gene regulation. BMC Genom. 2015, 16 (Suppl. 7). [Google Scholar] [CrossRef] [PubMed]
- Batmanov, K.; Wang, W.; Bjørås, M.; Delabie, J.; Wang, J. Integrative whole-genome sequence analysis reveals roles of regulatory mutations in BCL6 and BCL2 in follicular lymphoma. Sci. Rep. 2017, 7, 7040. [Google Scholar] [CrossRef] [PubMed]
- Miele, V.; Vaillant, C.; d’Aubenton-Carafa, Y.; Thermes, C.; Grange, T. DNA physical properties determine nucleosome occupancy from yeast to fly. Nucleic Acids Res. 2008, 36, 3746–3756. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.; Riley, T.; Liu, P.; Abe, N.; Gomez-Alcala, P.; Dror, I.; Zhou, T.; Rohs, R.; Honig, B.; Bussemaker, H.J.; et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 2011, 147, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Rohs, R.; West, S.M.; Sosinsky, A.; Liu, P.; Mann, R.S.; Honig, B. The role of DNA shape in protein-DNA recognition. Nature 2009, 461, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Shen, N.; Yang, L.; Abe, N.; Horton, J.; Mann, R.S.; Bussemaker, H.J.; Gordan, R.; Rohs, R. Quantitative modeling of transcription factor binding specificities using DNA shape. Proc. Natl. Acad. Sci. USA 2015, 112, 4654–4659. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Z.T.; Shiu, S.H.; Tsai, H.K. Contribution of sequence motif, chromatin state, and DNA structure features to predictive models of transcription factor binding in Yeast. PLoS Comput. Biol. 2015, 11, e1004418. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ramsey, S.A. A DNA shape-based regulatory score improves position-weight matrix-based recognition of transcription factor binding sites. Bioinformatics 2015, 31, 3445–3450. [Google Scholar] [CrossRef] [PubMed]
- Weirauch, M.T.; Cote, A.; Norel, R.; Annala, M.; Zhao, Y.; Riley, T.R.; Saez-Rodriguez, J.; Cokelaer, T.; Vedenko, A.; Talukder, S.; et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol. 2013, 31, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Friedel, M.; Nikolajewa, S.; Suhnel, J.; Wilhelm, T. DiProDB: A database for dinucleotide properties. Nucleic Acids Res. 2009, 37, D37–D40. [Google Scholar] [CrossRef] [PubMed]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Swinstead, E.E.; Miranda, T.B.; Paakinaho, V.; Baek, S.; Goldstein, I.; Hawkins, M.; Karpova, T.S.; Ball, D.; Mazza, D.; Lavis, L.D.; et al. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 2016, 165, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Fornes, O.; Mathelier, A.; Wasserman, W.W. Evaluating the impact of single nucleotide variants on transcription factor binding. Nucleic Acids Res. 2016, 44, 10106–10116. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Yang, L.; Lu, Y.; Dror, I.; Dantas Machado, A.C.; Ghane, T.; Di Felice, R.; Rohs, R. DNAshape: A method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013, 41, W56–W62. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. The effect of prior assumptions over the weights in BayesPI with application to study protein–DNA interactions from ChIP-based high-throughput data. BMC Bioinform. 2010, 11, 412. [Google Scholar] [CrossRef] [PubMed]
- Mackay, D. Bayesian Methods for Adaptive Models. Ph.D. Thesis, California Institute of Technology, Pasadena, CA, USA, 1991. [Google Scholar]
- Bewley, C.A.; Gronenborn, A.M.; Clore, G.M. Minor groove-binding architectural proteins: Structure, function, and DNA recognition. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 105–131. [Google Scholar] [CrossRef] [PubMed]
- Lazarovici, A.; Zhou, T.; Shafer, A.; Dantas Machado, A.C.; Riley, T.R.; Sandstrom, R.; Sabo, P.J.; Lu, Y.; Rohs, R.; Stamatoyannopoulos, J.A.; et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. USA 2013, 110, 6376–6381. [Google Scholar] [CrossRef] [PubMed]
- Segal, E.; Fondufe-Mittendorf, Y.; Chen, L.; Thastrom, A.; Field, Y.; Moore, I.K.; Wang, J.P.; Widom, J. A genomic code for nucleosome positioning. Nature 2006, 442, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Mathelier, A.; Fornes, O.; Arenillas, D.J.; Chen, C.Y.; Denay, G.; Lee, J.; Shi, W.; Shyr, C.; Tan, G.; Worsley-Hunt, R.; et al. JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016, 44, D110–D115. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lan, X.; Hsu, P.Y.; Hsu, H.K.; Huang, K.; Parvin, J.; Huang, T.H.; Jin, V.X. Genome-wide analysis uncovers high frequency, strong differential chromosomal interactions and their associated epigenetic patterns in E2-mediated gene regulation. BMC Genom. 2013, 14, 70. [Google Scholar] [CrossRef] [PubMed]
- Jozwik, K.M.; Carroll, J.S. Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer 2012, 12, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Naud, J.F.; McDuff, F.O.; Sauve, S.; Montagne, M.; Webb, B.A.; Smith, S.P.; Chabot, B.; Lavigne, P. Structural and thermodynamical characterization of the complete p21 gene product of Max. Biochemistry 2005, 44, 12746–12758. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Kawamoto, T.; Fujimoto, K.; Noshiro, M.; Honda, K.K.; Honma, S.; Honma, K.; Kato, Y. Functional analysis of the basic helix-loop-helix transcription factor DEC1 in circadian regulation. Interaction with BMAL1. Eur. J. Biochem. 2004, 271, 4409–4419. [Google Scholar] [CrossRef] [PubMed]
- Bolshoy, A. CC dinucleotides contribute to the bending of DNA in chromatin. Nat. Struct. Biol. 1995, 2, 446–448. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batmanov, K.; Wang, J. Predicting Variation of DNA Shape Preferences in Protein-DNA Interaction in Cancer Cells with a New Biophysical Model. Genes 2017, 8, 233. https://doi.org/10.3390/genes8090233
Batmanov K, Wang J. Predicting Variation of DNA Shape Preferences in Protein-DNA Interaction in Cancer Cells with a New Biophysical Model. Genes. 2017; 8(9):233. https://doi.org/10.3390/genes8090233
Chicago/Turabian StyleBatmanov, Kirill, and Junbai Wang. 2017. "Predicting Variation of DNA Shape Preferences in Protein-DNA Interaction in Cancer Cells with a New Biophysical Model" Genes 8, no. 9: 233. https://doi.org/10.3390/genes8090233




