Auxin Response Factor Genes Repertoire in Mulberry: Identification, and Structural, Functional and Evolutionary Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Transcriptome Assembly, Functional Annotation and Differential Expression Analyses
2.3. Identification and Phylogenetic Analyses of ARFs
2.4. Structural and Functional Analyses
2.5. Promoter Analysis, cis-Elements Identification and Their Enrichment Analysis
2.6. Stress and Hormonal Treatments and Relative Expression Analyses with qPCR
2.7. Interaction Networks of Mulberry ARF Genes
3. Results
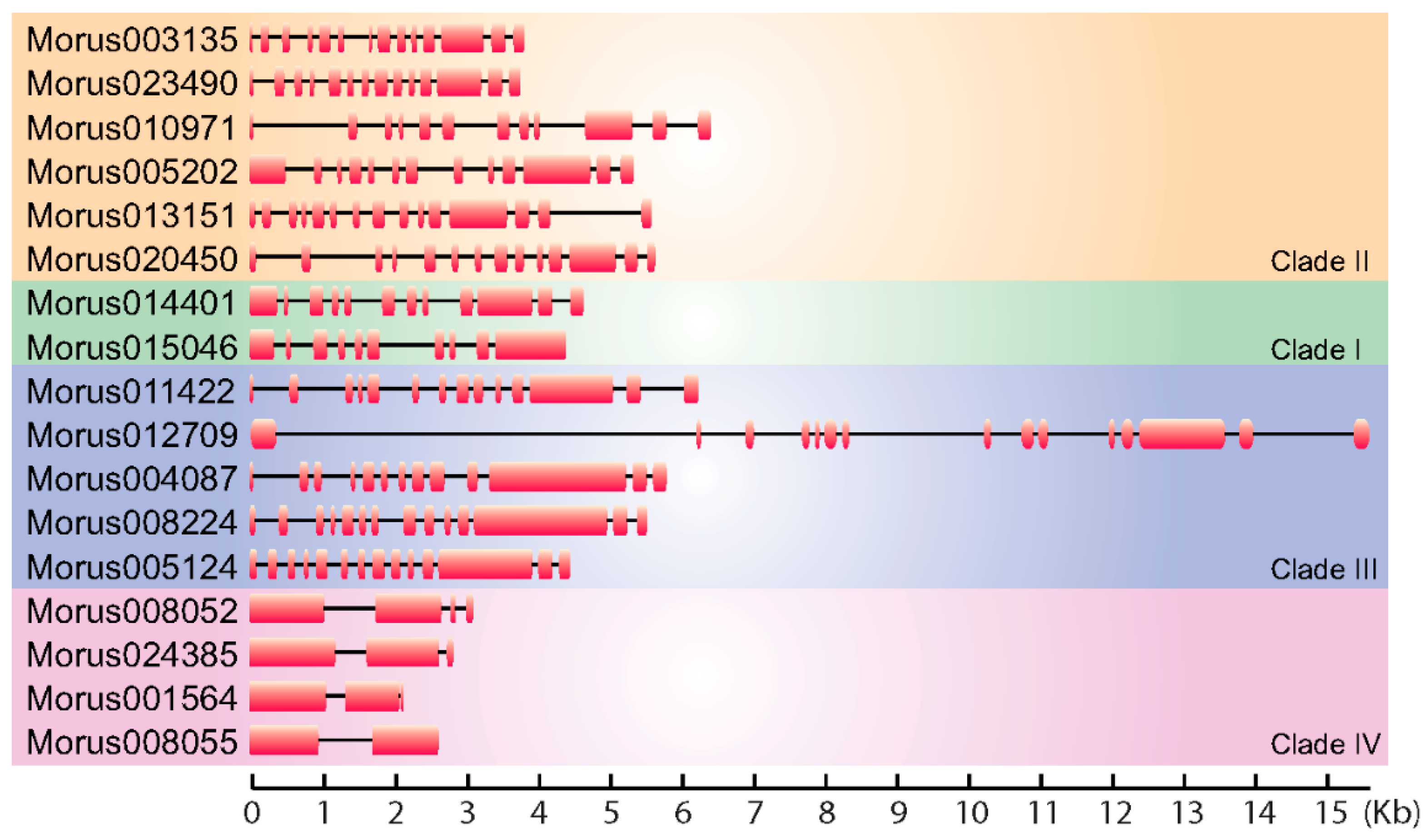
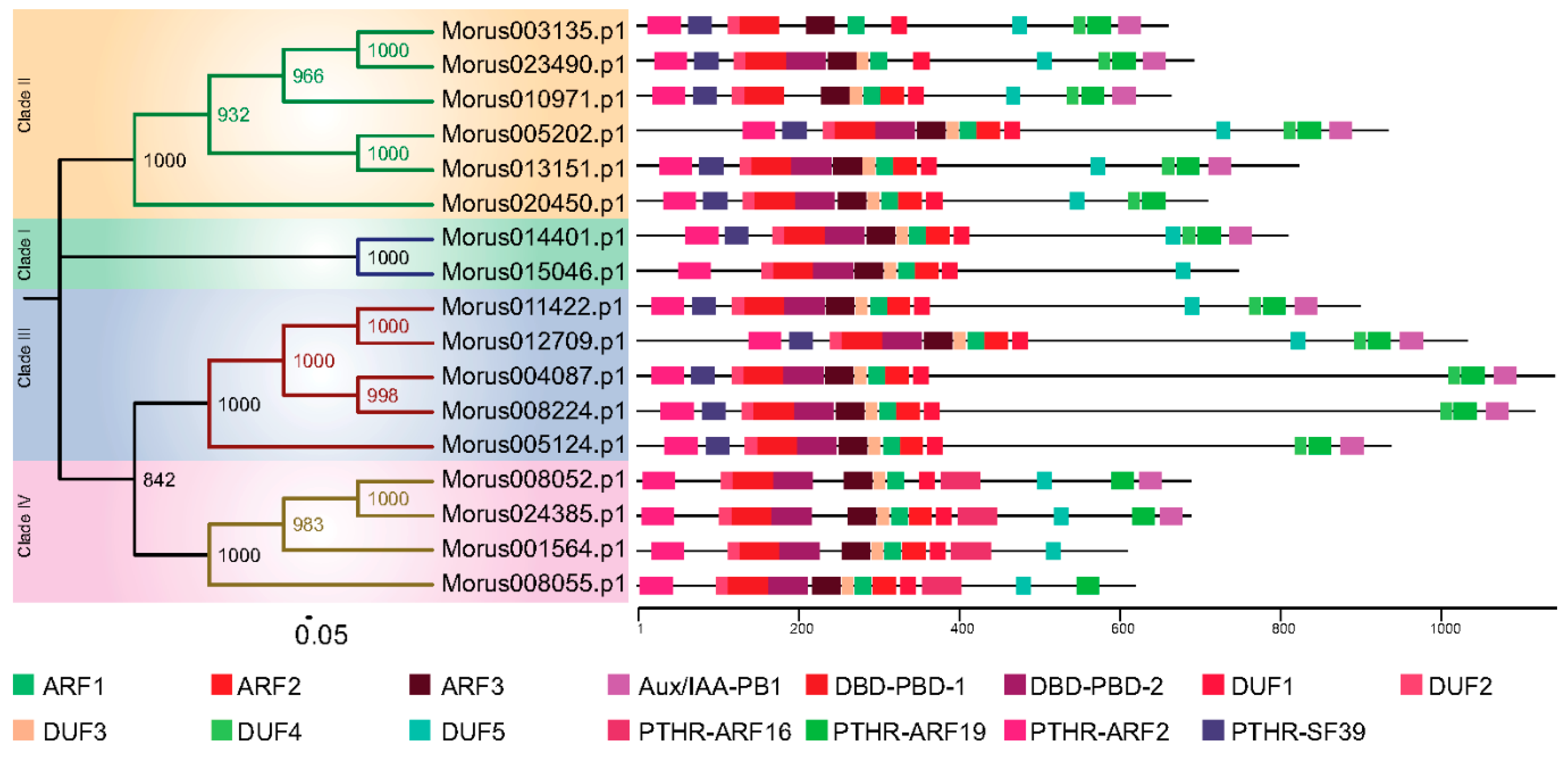
3.1. Structural Features of Mulberry ARFs Encoding Genes and Proteins
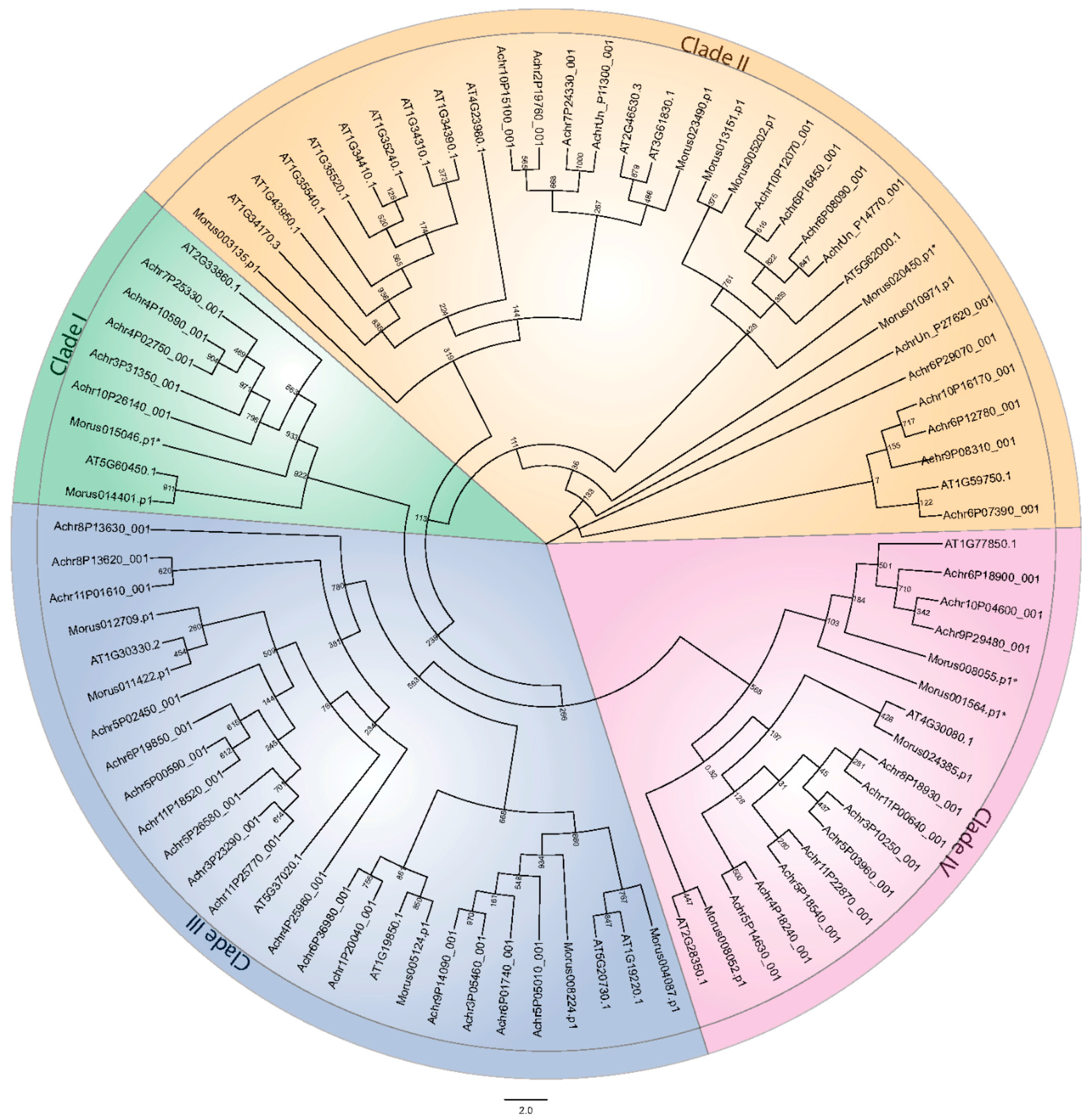
3.2. Phylogeny and Classification of Mulberry Auxin Response Factors
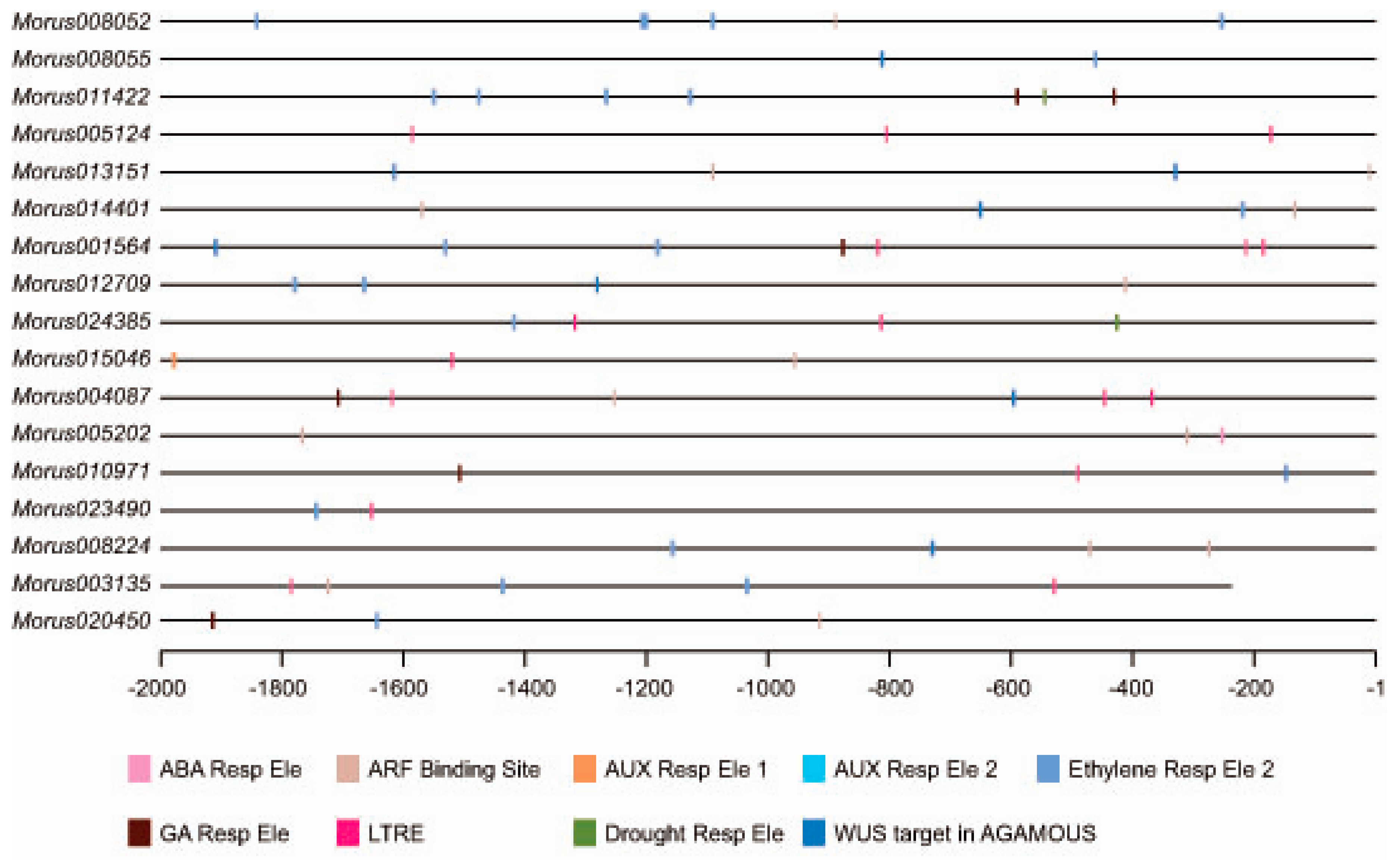
3.3. Distribution of cis Elements in 2 kb Upstream Region and Gene Ontology Enrichments Analysis
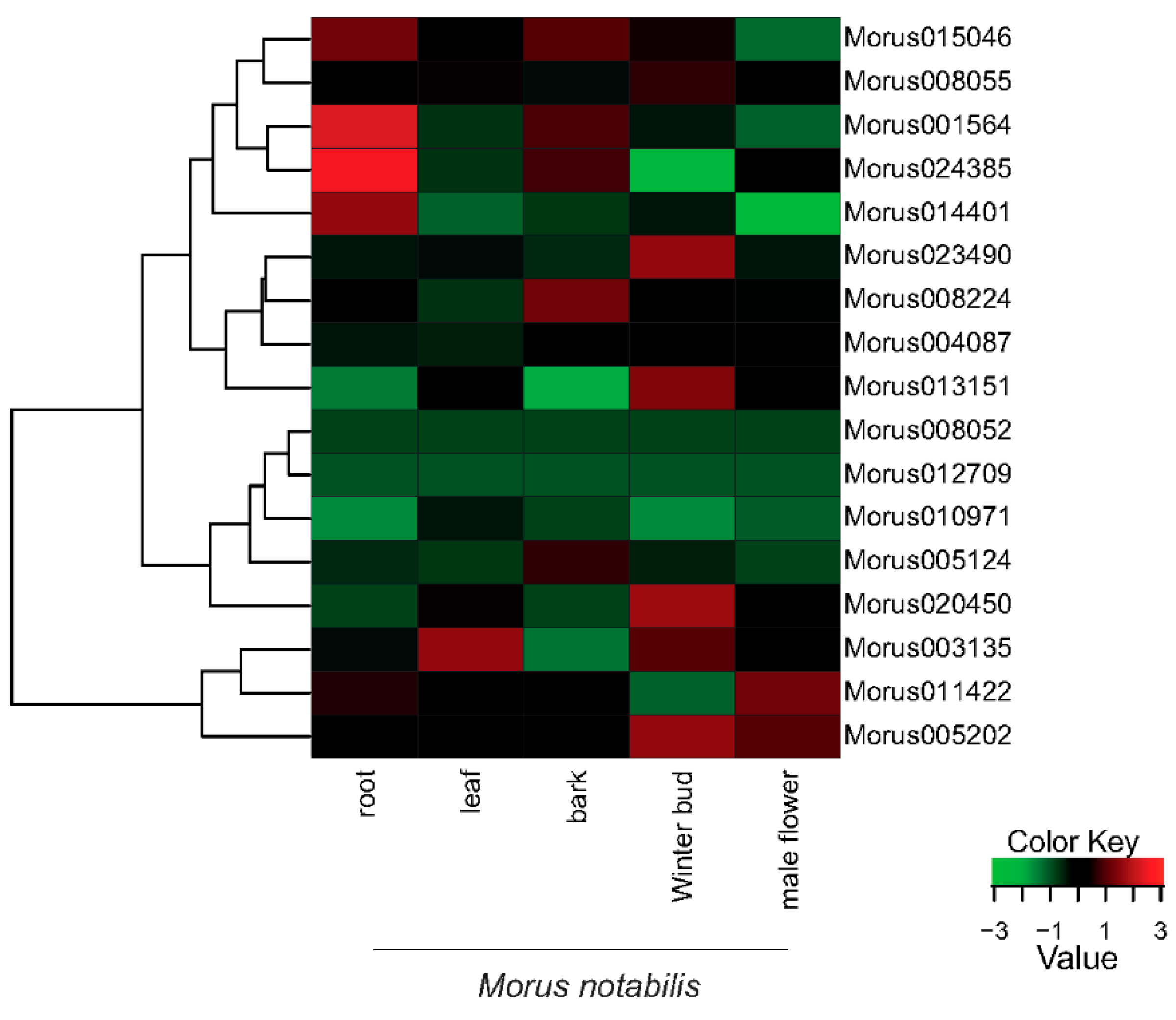
3.4. Expression Profiles of Auxin Response Factors in Developmental Stages of Morus Notabilis
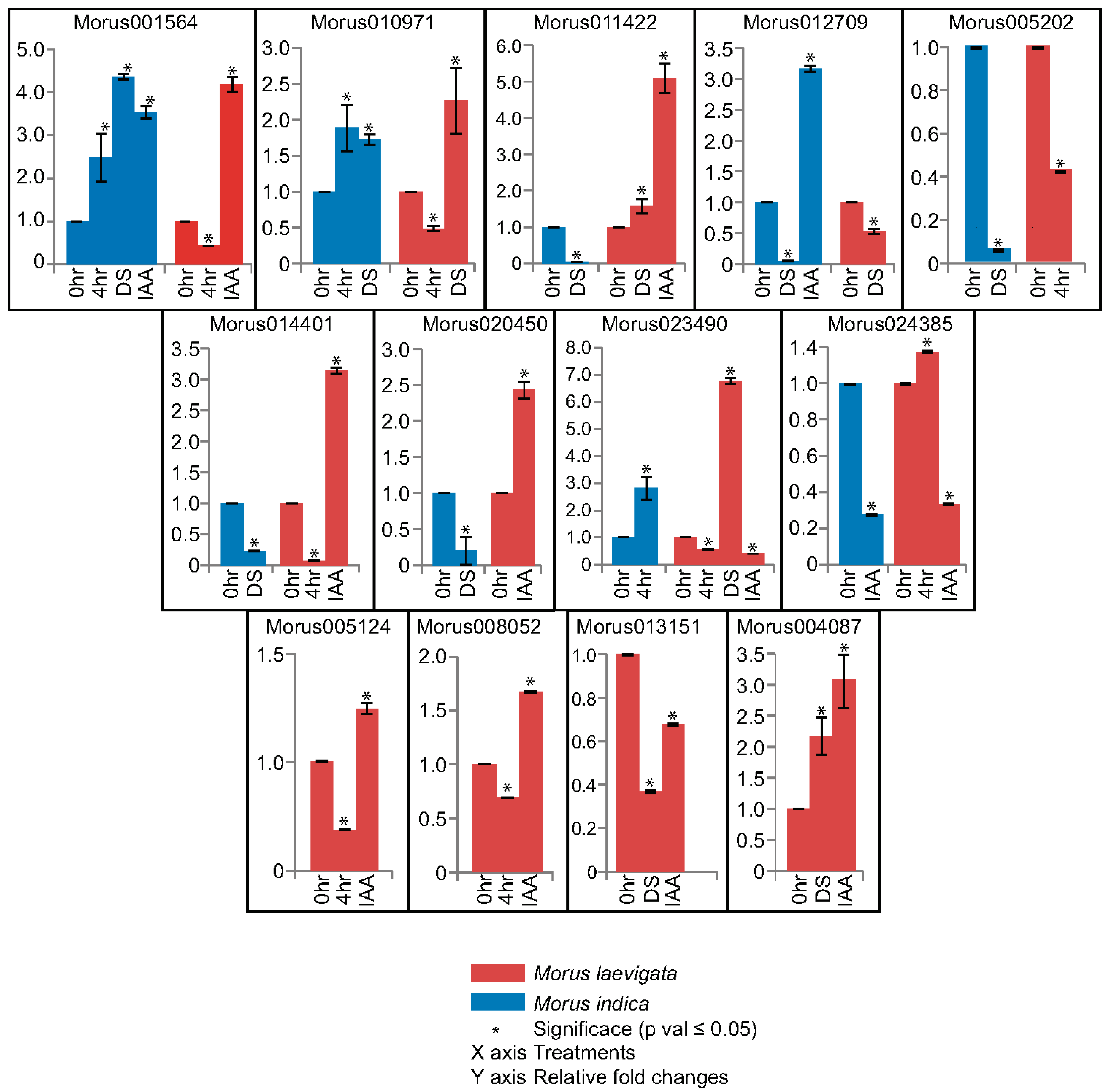
3.5. Differential Expression of ARF Genes in Three Wild Species of Morus
3.6. Genotype Specific Differential Expression of ARFs in Drought and Hormonal Treatment
3.7. Interaction Network of Mulberry ARFs
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author contributions
Conflicts of interest
References
- Lau, S.; Jürgens, G.; Smet, I.D. The evolving complexity of the auxin pathway. Plant Cell 2008, 20, 1738–1746. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K. The interaction and integration of auxin signaling components. Plant Cell Physiol. 2012, 53, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Finet, C.; Fourquin, C.; Vinauger, M.; Berne-Dedieu, A.; Chambrier, P.; Paindavoine, S.; Scutt, C.P. Parallel structural evolution of auxin response factors in the angiosperms: ARF evolution in angiosperms. Plant J. 2010, 63, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, U.C.; Difazio, S.P.; Brunner, A.M.; Tuskan, G.A. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Soler, M.; Mila, I.; San Clemente, H.; Savelli, B.; Dunand, C.; Paiva, J.A.P.; Myburg, A.A.; Bouzayen, M.; Grima-Pettenati, J.; et al. Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PLoS ONE 2014, 9, e108906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Tyagi, A.K.; Sharma, A.K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genom. 2011, 285, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dai, X.; Zhao, Y. A Role for Auxin Response Factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol. 2006, 140, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Omori, Y.; Nagasawa, H. Dietary sterol preference in the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 2006, 70, 3094–3098. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, F.; Rodella, R.A. Características morfo-anatômicas e bromatológicas de folhas de amoreira em relação às preferências do bicho-da-seda. Pesqui. Agropecuária Bras. 2006, 41, 195–203. [Google Scholar] [CrossRef]
- Barathi, P.; Sundar, D.; Reddy, A.R. Changes in mulberry leaf metabolism in response to water stress. Biol. Plant. 2001, 44, 83–87. [Google Scholar] [CrossRef]
- Chatterjee, S.N.; Nagaraja, G.M.; Srivastava, P.P.; Naik, G. Morphological and molecular variation of Morus laevigata in India. Genetica 2004, 121, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Tikader, A.; Thangavelu, K. Breeding performance of some wild mulberry (Morus spp.). Proc. Nat. Conf. Rec. Trends Plant Sci. Res. 2002, 106–111. [Google Scholar]
- Babu, A.M.; Kumar, J.S.; Kumar, V.; Sarkar, A.; Datta, R.K. Tropic failure of Phyllactinia corylea contributes to the mildew resistance of mulberry genotypes. Mycopathologia 2002, 156, 207–213. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Zhang, C.; Qi, X.; Zhao, S.; Tao, Y.; Yang, G.; Lee, T.-H.; Wang, X.; Cai, Q.; Li, D. Draft genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 2013, 4, 2445. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Qi, X.; Zeng, Q.; Xiang, Z.; He, N. MorusDB: A resource for mulberry genomics and genome biology. Database 2014. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, V.K.; Khurana, P. Membrane intrinsic proteins repertoire of Morus notabilis and their expression profiles in different species. Plant Physiol. Biochem. 2016. [Google Scholar] [CrossRef]
- Saeed, B.; Baranwal, V.K.; Khurana, P. Comparative transcriptomics and comprehensive marker resource development in mulberry. BMC Genom. 2016, 17, 98. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, V.K.; Khurana, P. Genome-wide analysis, expression dynamics and varietal comparison of NAC gene family at various developmental stages in Morus notabilis. Mol. Genet. Genom. 2016, 291, 1305–1317. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 16 August 2014).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-W.; Gan, R.-C.R.; Wu, T.H.; Huang, P.-J.; Lee, C.-Y.; Chen, Y.-Y.M.; Chen, C.-C.; Tang, P. FastAnnotator-an efficient transcript annotation web tool. BMC Genom. 2012, 13, S9. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- D’Hont, A.; Denoeud, F.; Aury, J.-M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E.; Block, D.; Boulez, K.; Brenner, S.E.; Chervitz, S.A.; Dagdigian, C.; Fuellen, G.; Gilbert, J.G.R.; Korf, I.; Lapp, H.; et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002, 12, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Higo, H. PLACE: A database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998, 26, 358–359. [Google Scholar] [CrossRef] [PubMed]
- Zambelli, F.; Pesole, G.; Pavesi, G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009, 37, W247–W252. [Google Scholar] [CrossRef] [PubMed]
- Mathelier, A.; Zhao, X.; Zhang, A.W.; Parcy, F.; Worsley-Hunt, R.; Arenillas, D.J.; Buchman, S.; Chen, C.; Chou, A.; Ienasescu, H.; et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014, 42, D142–D147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baranwal, V.K.; Negi, N.; Khurana, P. Genome-wide Identification and Structural, Functional and Evolutionary Analysis of WRKY Components of Mulberry. Sci. Rep. 2016, 6, 30794. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Hu, W.; Zuo, J.; Hou, X.; Yan, Y.; Wei, Y.; Liu, J.; Li, M.; Xu, B.; Jin, Z. The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-B.; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Dimerization and DNA binding of auxin response factors. Plant J. 1999, 19, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Itzhaki, H.; Maxson, J.M.; Woodson, W.R. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc. Natl. Acad. Sci. USA 1994, 91, 8925–8929. [Google Scholar] [CrossRef] [PubMed]
- Kizis, D.; Pagès, M. Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J. 2002, 30, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.L.; Hamaguchi, L.; Busch, M.A.; Weigel, D. Regulatory Elements of the Floral Homeotic Gene AGAMOUS Identified by Phylogenetic Footprinting and Shadowing. Plant Cell 2003, 15, 1296–1309. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Mizunashi, K.; Tanaka, M.; Kaminuma, E.; Nguyen, A.H.; Nakajima, M.; Kim, J.-M.; Nguyen, D.V.; Toyoda, T.; Seki, M. tasiRNA-ARF pathway moderates floral architecture in Arabidopsis plants subjected to drought stress. BioMed Res. Int. 2014, 2014, 303451. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.; Wolters-Arts, M.; Schimmel, B.C.J.; Stultiens, C.L.M.; de Groot, P.F.M.; Powers, S.J.; Tikunov, Y.M.; Bovy, A.G.; Mariani, C.; Vriezen, W.H.; et al. Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot. 2015, 66, 3405–3416. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. AUXIN RESPONSE FACTOR 7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 2005, 17, 1979–1993. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [PubMed]
- Naik, M.V.G.; Thumilan, B.M.; Roy, B.; Dandin, S.B. Assessment of genetic diversity and interrelationship among wild mulberry (Morus laevigata and Morus Serrata) collections of India through DNA marker analysis. In Proceedings of the Second National Plant Breeding Congress on Plant Breeding in Post Genomics Era, Coimbatore, India, 1–3 March 2006; Indian Society of Plant Breeders: New Delhi, India, 2006; pp. 302–308. [Google Scholar]
- Krogan, N.T.; Berleth, T. The identification and characterization of specific ARF-Aux/IAA regulatory modules in plant growth and development. Plant Signal. Behav. 2015, 10, e992748. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ma, B.; Qi, X.; Guo, Q.; Wang, X.; Zeng, Q.; He, N. Identification and characterization of genes involved in the jasmonate biosynthetic and signaling pathways in mulberry Morus notabilis: Analysis of mulberry jasmonate-related genes. J. Integr. Plant Biol. 2014, 56, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Yuan, C.; Li, H.; Lin, W.; Yang, Y.; Shen, C.; Zheng, X. Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica papaya L.). BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, F.; Cheng, L.; Kong, F.; Peng, Z.; Liu, S.; Yu, X.; Lu, G. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 2011, 30, 2059. [Google Scholar] [CrossRef] [PubMed]
- Baba, K.; Karlberg, A.; Schmidt, J.; Schrader, J.; Hvidsten, T.R.; Bako, L.; Bhalerao, R.P. Activity-dormancy transition in the cambial meristem involves stage-specific modulation of auxin response in hybrid aspen. Proc. Natl. Acad. Sci. USA 2011, 108, 3418–3423. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baranwal, V.K.; Negi, N.; Khurana, P. Auxin Response Factor Genes Repertoire in Mulberry: Identification, and Structural, Functional and Evolutionary Analyses. Genes 2017, 8, 202. https://doi.org/10.3390/genes8090202
Baranwal VK, Negi N, Khurana P. Auxin Response Factor Genes Repertoire in Mulberry: Identification, and Structural, Functional and Evolutionary Analyses. Genes. 2017; 8(9):202. https://doi.org/10.3390/genes8090202
Chicago/Turabian StyleBaranwal, Vinay Kumar, Nisha Negi, and Paramjit Khurana. 2017. "Auxin Response Factor Genes Repertoire in Mulberry: Identification, and Structural, Functional and Evolutionary Analyses" Genes 8, no. 9: 202. https://doi.org/10.3390/genes8090202



