HiSeeker: Detecting High-Order SNP Interactions Based on Pairwise SNP Combinations
Abstract
:1. Introduction
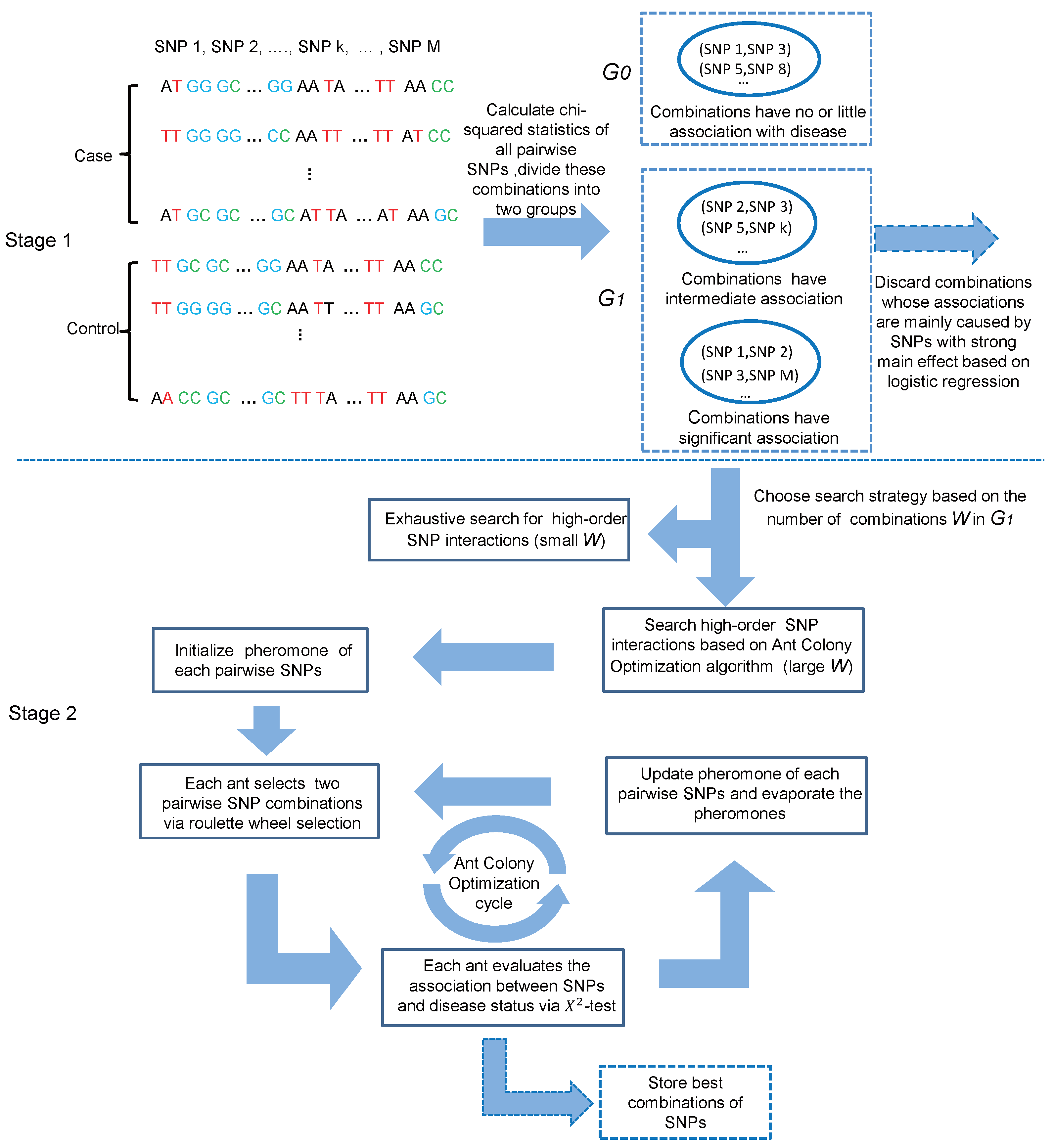
2. Materials and Methods
2.1. Stage I: Valid Two-Locus Combination Candidate Selection
2.1.1. Two-Locus Combination Filtering
- For given genotype data with M SNPs, to measure the association between the combination ( and disease, a contingency table like Table 1 is firstly constructed, then chi-squared statistic is calculated as:where follows a chi-squared distribution with 8 degrees of freedom. The chi-squared statistics of all two-locus combinations in G are denoted as .
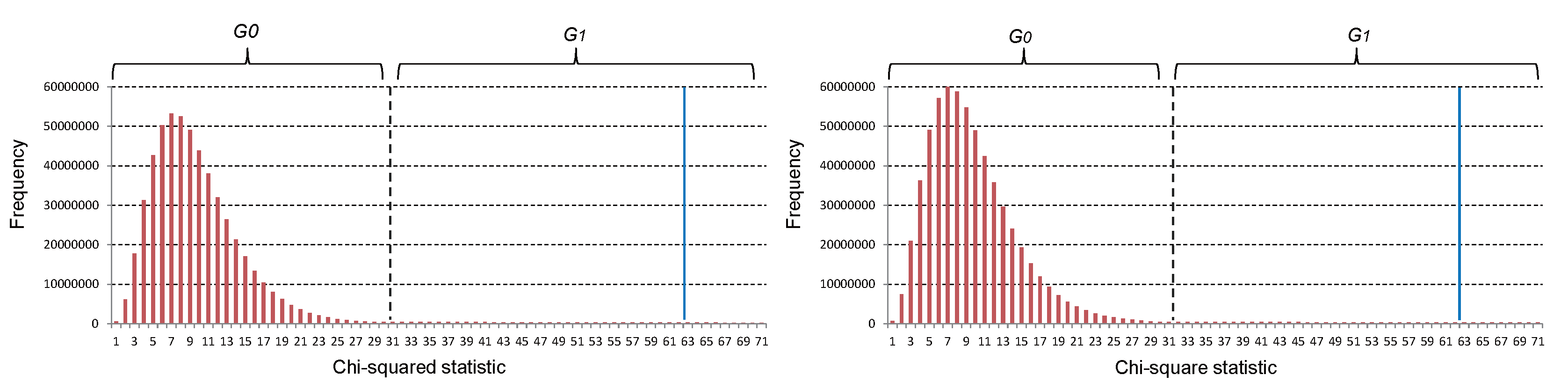
- To obtain combinations having significant or intermediate association with disease, we firstly distinguish significant combinations from G that pass the Bonferroni correction. A combination with a statistic is placed into , where denotes the corresponding chi-squared statistic of the Bonferroni-corrected significance level . Given a preset significance level , is calculated as:
- To obtain combinations having intermediate association with disease, a significance level is defined as:where is a scale factor that adjusts the number of combinations in , which is retained for following analysis. The combinations with the chi-squared statistic between and have intermediate association with disease; they are also placed into . All of the other combinations are placed into .
2.1.2. Candidate Combinations Screening
- For a two-locus combination (), HiSeeker first fits a full logistic regression model to measure the full association between () and disease status Y (1 for case and 0 for control) as follows:where and are the main effects for SNP and , respectively, and represents the interaction effect. Then, the Newton–Raphson method is utilized to iteratively optimize the corresponding maximum likelihood value of Equation (4).
- For a two-locus combination, if both and have strong main effects, a logistic regression model defined by Equation (5) is fitted to measure the additive main effects of them as:If only (or ) has a strong main effect, a logistic regression model defined by Equation (6) is used to measure the main effect in that combination as:Then, the Newton–Raphson method is utilized again to iteratively optimize the corresponding maximum likelihood value of Equation (5) or Equation (6).
- HiSeeker calculates the deviation D of each two-locus combination in as follows:where D follows a chi-squared distribution with degree of freedom . if both and have strong marginal effect; if only (or ) has strong main effect.
2.2. Stage 2: High-Order SNP Interaction Detection
2.2.1. Exhaustive Search Strategy for Small Candidate Set (Small W)
2.2.2. Ant Colony Optimization Strategy for Large Candidate Set (Large W)
- (i)
- Initialization: the pheromone value of each two-locus combination is initialized as a fixed value , which means that the association between a combination and disease is treated with equal possibility. To identify possible candidate combinations to assemble the high-order SNP interaction sets, ACO iteratively selects and evaluates SNP combinations from W candidates via the following Step ii to Step iv, until a preset number of iterations is reached.
- (ii)
- Combination selection: ACO introduces n operators called ants to select SNP combinations. n is set based on the candidate size W (). In each iteration, an ant chooses d () combinations as its targeted two-locus combination set. d is set according to the order number needed by users. To detect three-locus interactions, d is initially set as 2. The probability for an ant x () selecting a two-locus combination based on roulette wheel selection can be defined as:where is the pheromone value of and is the prior information on . and are parameters to determine the weight of pheromone value and the weight of prior information on each combination, respectively. Here, , and are set to 1, indicating that each combination is treated equally before the optimization phase.
- (iii)
- Evaluation on the selected combinations: the statistic of the chi-squared test is applied as the fitness function. In each iteration, two selected combinations of each ant are merged into a new combination . The fitness of is calculated using Equation (1) and denoted as . If the number of SNPs in is 3 (or 4), follows a chi-squared distribution with degree of freedom (or ). Given the same significance level , the Bonferroni-corrected significance level is (or ) for the three-locus (or four-locus) combination. The corresponding chi-square statistic is (or ). is about two-times . Thus, to avoid the loss of significant three-locus combinations, is multiplied with a scale factor when the number of SNPs in is 3. For HiSeeker, . In each iteration, the merged SNP combinations with the highest chi-squared statistics are stored.
- (iv)
- Pheromone update: in each iteration, after the selected d two-locus combinations of each ant have been evaluated, the corresponding pheromone of each two-locus combination in an ant is updated as:where is the evaporating coefficient and is the changing pheromone value of the i-th two-locus combination , which equals 0.01 of this ant. This update process is repeated for all ants.
3. Results
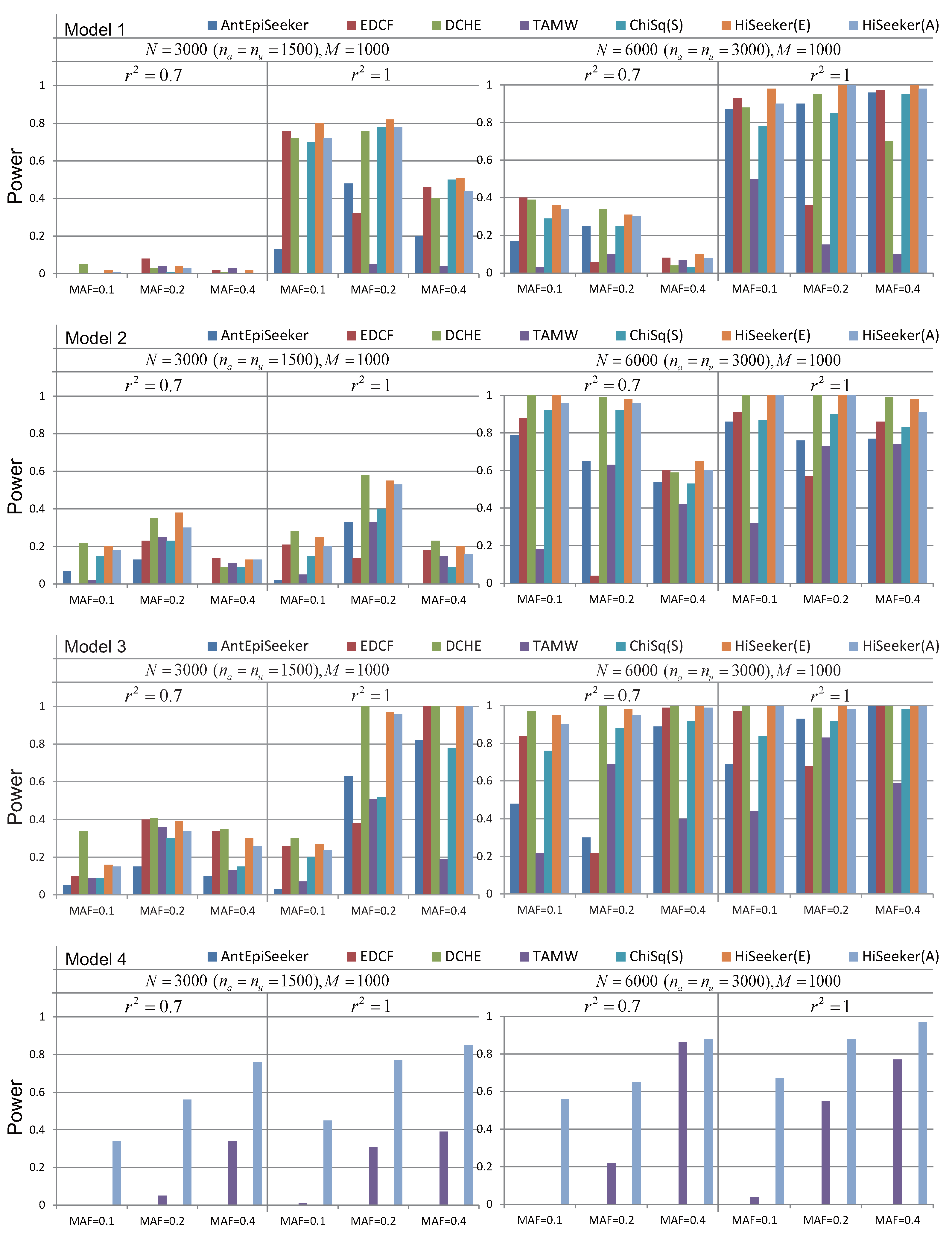
3.1. Experiments on Simulated Datasets
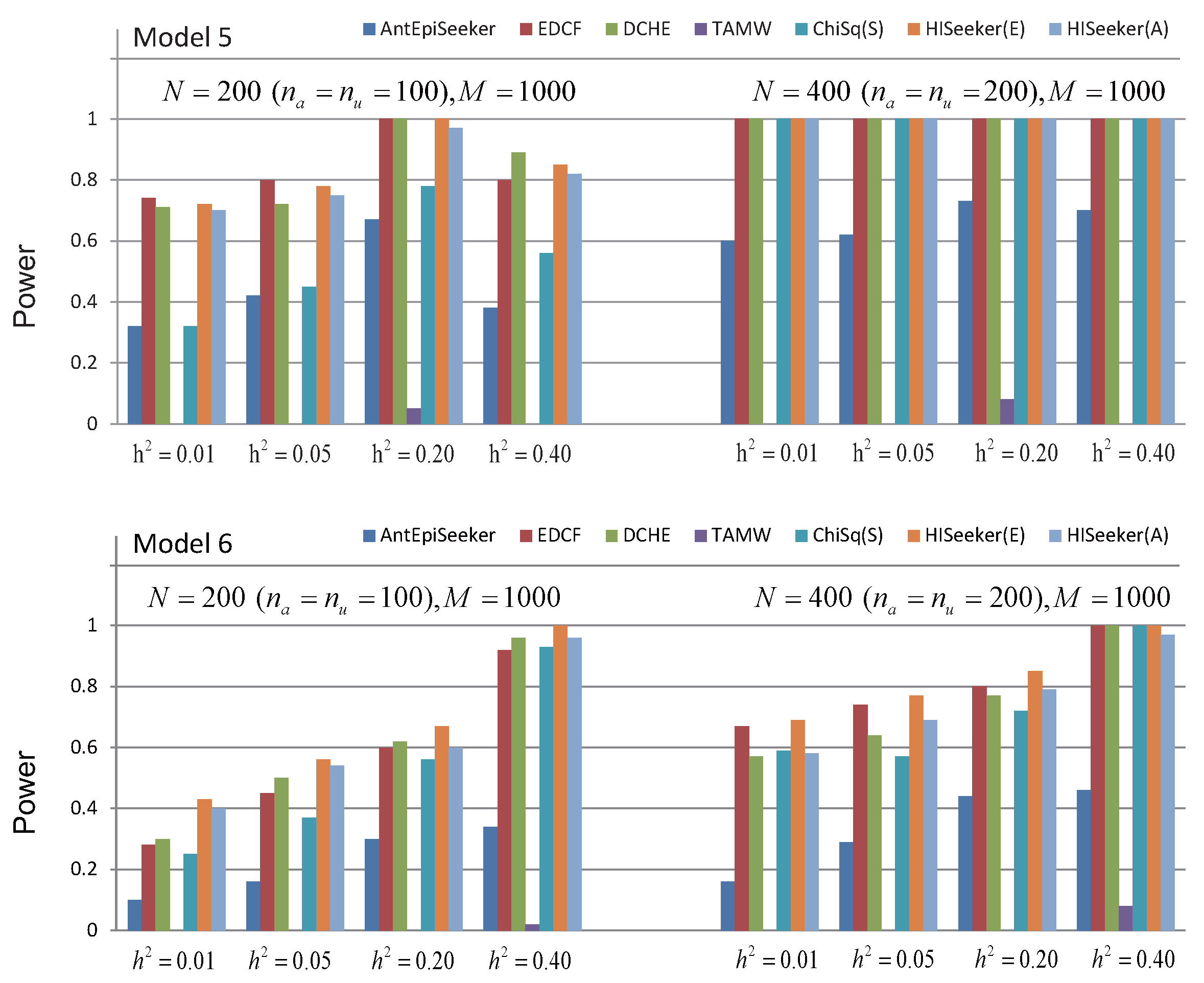
3.1.1. Case 1: Disease Loci with Marginal Effects
3.1.2. Case 2: Disease Loci without Marginal Effects
3.2. Experiments on Real Datasets
3.2.1. Experiments on BC Data
3.2.2. Experiments on CD Data
3.3. Parameter Setting
- of each two-locus combination is always set to 100, which means that we treat the association between each combination and disease with equal probability.
- For K-locus interaction detection, d should be set bigger than . In the simulation study, we set for three-locus interaction detection and set for six-locus interaction detection.
- ranges from 0.01 to 0.1 according to the number of candidate two-locus combinations W. A large should be adopted for a small W. In the simulation study, we set = 0.05. In the real study, is set to 0.01.
- Both n and MaxIter are determined by W. We set MaxIter = 0.1 W. n ranges from 500 to 5000. A large W prefers large n and MaxIter.
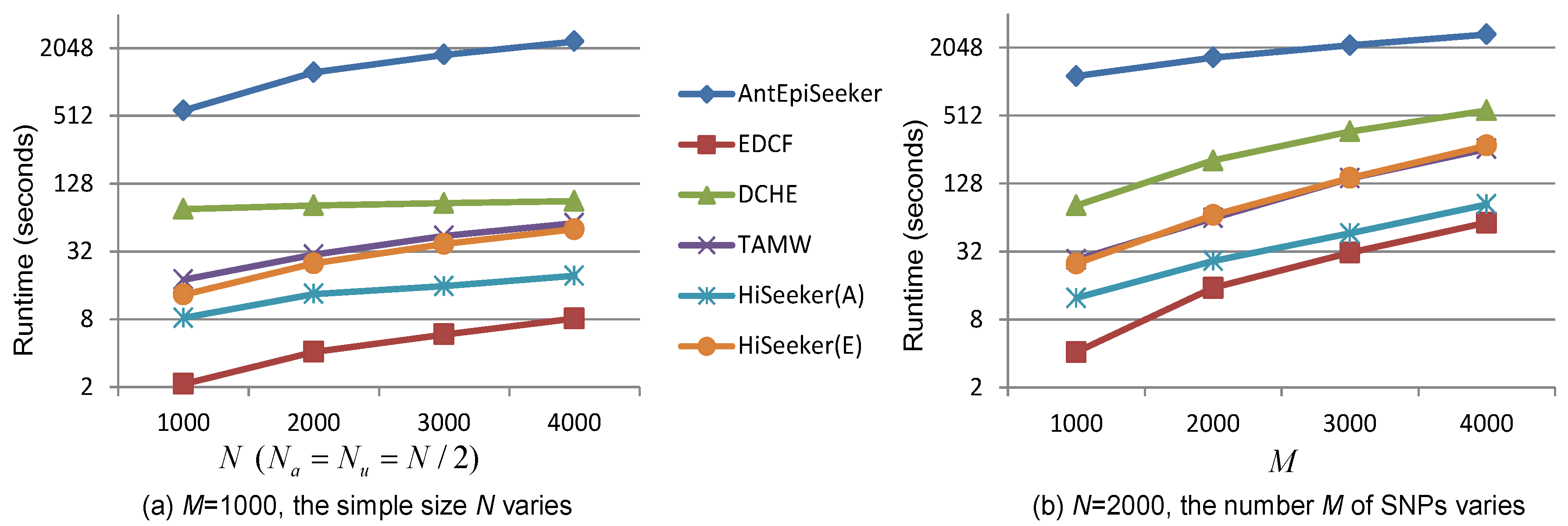
3.4. Runtime Analysis
4. Conclusions
- HiSeeker flexibly screens two-locus combinations with strong or intermediate association with disease phenotype. This flexibility enables it to detect more high-order interactions, whose decomposed pairwise interactions are not significant.
- HiSeeker is not sensitive to the marginal effects of individual SNPs; since it makes use of the likelihood ratio test based on logistic regression to filter out the two-locus combinations, whose associations with disease are mainly caused by the SNPs with strong main effects.
- HiSeeker provides two alternative search strategies for datasets with different scales, and it enables detecting high-order interaction on large GWAS data without exhaustive enumeration.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Welter, D.; MacArthur, J.; Morales, J.; Burdett, T.; Hall, P.; Junkins, H.; Klemm, A.; Flicek, P.; Manolio, T.; Hindorff, L.; et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014, 42, D1001–D1006. [Google Scholar] [CrossRef] [PubMed]
- Cantor, R.M.; Lange, K.; Sinsheimer, J.S. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am. J. Hum. Genet. 2010, 86, 6–22. [Google Scholar] [CrossRef] [PubMed]
- Kraft, P.; Hunter, D.J. Genetic risk prediction—Are we there yet? N. Engl. J. Med. 2009, 360, 1701–1703. [Google Scholar] [CrossRef] [PubMed]
- Witte, J.S.; Visscher, P.M.; Wray, N.R. The contribution of genetic variants to disease depends on the ruler. Nat. Rev. Genet. 2014, 15, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.H.; Hemani, G.; Haley, C.S. Detecting epistasis in human complex traits. Nat. Rev. Genet. 2014, 15, 722–733. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.Y.; Wang, Y.T.; Yu, Y.W.; Chung, R.H. An efficient gene—Gene interaction test for genome-wide association studies in trio families. Bioinformatics 2016, 32, 1848–1855. [Google Scholar] [CrossRef] [PubMed]
- Fish, A.E.; Capra, J.A.; Bush, W.S. Are Interactions between cis-Regulatory Variants Evidence for Biological Epistasis or Statistical Artifacts? Am. J. Hum. Genet. 2016, 99, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Phillips, P.C. Epistasis—The essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008, 9, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Marchini, J.; Donnelly, P.; Cardon, L.R. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 2005, 37, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Van Steen, K. Travelling the world of gene—Gene interactions. Brief. Bioinform. 2012, 13, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Cordell, H.J. Detecting gene—Gene interactions that underlie human diseases. Nat. Rev. Genet. 2009, 10, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Yang, C.; Yang, Q.; Xue, H.; Fan, X.; Tang, N.L.; Yu, W. BOOST: A fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am. J. Hum. Genet. 2010, 87, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, S.; Zou, F.; Wang, W. TEAM: Efficient two-locus epistasis tests in human genome-wide association study. Bioinformatics 2010, 26, i217–i227. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.B.; Ehrenreich, I.M. Higher-order genetic interactions and their contribution to complex traits. Trends Genet. 2015, 31, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, D.M.; Lan, Y.; Wylie, C.S.; Heckendorn, R.B. Should evolutionary geneticists worry about higher-order epistasis? Curr. Opin. Genet. Dev. 2013, 23, 700–707. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Qian, W.; Wang, Z.; Li, Y.; Zhang, J. Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks. Nat. Genet. 2010, 42, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ding, J.; Effgen, S.; Turck, F.; Koornneef, M. Multiple loci and genetic interactions involving flowering time genes regulate stem branching among natural variants of Arabidopsis. New Phytol. 2013, 199, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.L.; Hu, T.; Wejse, C.; Sirugo, G.; Williams, S.M.; Moore, J.H. Multifactor dimensionality reduction reveals a three-locus epistatic interaction associated with susceptibility to pulmonary tuberculosis. BioData Min. 2013, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Hahn, L.W.; Roodi, N.; Bailey, L.R.; Dupont, W.D.; Parl, F.F.; Moore, J.H. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001, 69, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Chen, Y.; Kiralis, J.W.; Collins, R.L.; Wejse, C.; Sirugo, G.; Williams, S.M.; Moore, J.H. An information-gain approach to detecting three-way epistatic interactions in genetic association studies. J. Am. Med. Inform. Assoc. 2013, 20, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Goudey, B.; Abedini, M.; Hopper, J.L.; Inouye, M.; Makalic, E.; Schmidt, D.F.; Wagner, J.; Zhou, Z.; Zobel, J.; Reumann, M. High performance computing enabling exhaustive analysis of higher order single nucleotide polymorphism interaction in Genome Wide Association Studies. Health Inf. Sci. Syst. 2015, 3, S3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Robbins, K.; Rekaya, R. AntEpiSeeker: Detecting epistatic interactions for case-control studies using a two-stage ant colony optimization algorithm. BMC Res. Notes 2010, 3, 117. [Google Scholar] [CrossRef] [PubMed]
- Aflakparast, M.; Salimi, H.; Gerami, A.; Dubé, M.; Visweswaran, S.; Masoudi-Nejad, A. Cuckoo search epistasis: A new method for exploring significant genetic interactions. Heredity 2014, 112, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Joshi, T.; Valliyodan, B.; Shi, H.; Liang, Y.; Nguyen, H.T.; Zhang, J.; Xu, D. A Bayesian model for detection of high-order interactions among genetic variants in genome-wide association studies. BMC Genom. 2015, 16, 1011. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Lu, Q. GWGGI: Software for genome-wide gene-gene interaction analysis. BMC Genet. 2014, 15, 101. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Wei, C.; Ye, C.; Li, M.; Elston, R.C. A likelihood ratio-based Mann-Whitney approach finds novel replicable joint gene action for type 2 Diabetes. Genet. Epidemiol. 2012, 36, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Schaid, D.J.; Lu, Q. Trees assembling Mann-Whitney approach for detecting genome-wide joint association among low-marginal-effect loci. Genet. Epidemiol. 2013, 37, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Li, J.; Jiang, T. Detecting genome-wide epistases based on the clustering of relatively frequent items. Bioinformatics 2012, 28, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Meng, Y.; Yu, N.; Pan, Y. Cloud computing for detecting high-order genome-wide epistatic interaction via dynamic clustering. BMC Bioinform. 2014, 15, 102. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Sun, Y.; Liu, J.X.; Xia, J.; Zhang, J.; Zheng, C.H. CINOEDV: A co-information based method for detecting and visualizing n-order epistatic interactions. BMC Bioinform. 2016, 17, 214. [Google Scholar] [CrossRef] [PubMed]
- Dubois, P.C.; Trynka, G.; Franke, L.; Hunt, K.A.; Romanos, J.; Curtotti, A.; Zhernakova, A.; Heap, G.A.; Ádány, R.; Aromaa, A.; et al. Multiple common variants for Celiac disease influencing immune gene expression. Nat. Genet. 2010, 42, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Goudey, B.; Rawlinson, D.; Wang, Q.; Shi, F.; Ferra, H.; Campbell, R.M.; Stern, L.; Inouye, M.T.; Ong, C.S.; Kowalczyk, A. GWIS-model-free, fast and exhaustive search for epistatic interactions in case-control GWAS. BMC Genom. 2013, 14, S10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zou, F.; Wang, W. FastChi: An efficient algorithm for analyzing gene-gene interactions. In Pacific Symposium on Biocomputing; NIH Public Access: Honolulu, HI, USA, 2009; p. 528. [Google Scholar]
- Jing, P.J.; Shen, H.B. MACOED: A multi-objective ant colony optimization algorithm for SNP epistasis detection in genome-wide association studies. Bioinformatics 2015, 31, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, H.; Chen, S.; Chen, X.; Zhang, Z.; Zhu, Z.; Qin, X.; Hu, L.; Zhu, J.; Zhao, G.P.; et al. Genome-wide interaction-based association analysis identified multiple new susceptibility loci for common diseases. PLoS Genet. 2011, 7, e1001338. [Google Scholar] [CrossRef] [PubMed]
- Hosmer, D.W., Jr.; Lemeshow, S. Applied Logistic Regression; John Wiley & Sons: Hoboken, NY, USA, 2004. [Google Scholar]
- Dorigo, M.; Gambardella, L.M. Ant colonies for the travelling salesman problem. BioSystems 1997, 43, 73–81. [Google Scholar] [CrossRef]
- Greene, C.S.; White, B.C.; Moore, J.H. Ant colony optimization for genome-wide genetic analysis. In International Conference on Ant Colony Optimization and Swarm Intelligence; Springer: Berlin, Germany, 2008; pp. 37–47. [Google Scholar]
- Sapin, E.; Keedwell, E.; Frayling, T. An ant aolony optimization and tabu list approach to the detection of gene-gene interactions in genome-wide association studies. IEEE Comput. Intell. Mag. 2015, 10, 54–65. [Google Scholar] [CrossRef]
- Tang, W.; Wu, X.; Jiang, R.; Li, Y. Epistatic module detection for case-control studies: A Bayesian model with a Gibbs sampling strategy. PLoS Genet. 2009, 5, e1000464. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Yang, C.; Yang, Q.; Xue, H.; Tang, N.L.; Yu, W. Predictive rule inference for epistatic interaction detection in genome-wide association studies. Bioinformatics 2010, 26, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, J.S. Bayesian inference of epistatic interactions in case-control studies. Nat. Genet. 2007, 39, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Culverhouse, R.; Suarez, B.K.; Lin, J.; Reich, T. A perspective on epistasis: limits of models displaying no main effect. Am. J. Hum. Genet. 2002, 70, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.H.; Urbanowicz, R.J.; Andrews, P. GAMETES 2.0: Expanding the complex model and data simulation software to generate heterogeneous datasets, custommodels, and quantitative traits. Genet. Epidemiol. 2015, 39, 570. [Google Scholar]
- Michailidou, K.; Hall, P.; Gonzalez-Neira, A.; Ghoussaini, M.; Dennis, J.; Milne, R.L.; Schmidt, M.K.; Chang-Claude, J.; Bojesen, S.E.; Bolla, M.K.; et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013, 45, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Burton, P.R.; Clayton, D.G.; Cardon, L.R.; Craddock, N.; Deloukas, P.; Duncanson, A.; Kwiatkowski, D.P.; McCarthy, M.I.; Ouwehand, W.H.; Samani, N.J.; et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 2007, 39, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, S.A.; Settasatian, C.; Crawford, J.; Lower, K.M.; McCallum, B.; Seshadri, R.; Cornelisse, C.J.; Moerland, E.W.; Cleton-Jansen, A.M.; Tipping, A.J.; et al. Characterization and screening for mutations of the growth arrest-specific 11 (GAS11) and C16orf3 genes at 16q24. 3 in breast cancer. Genomics 1998, 52, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Geng, S.; Jin, F.; Liu, J.; Qu, C.; Chen, B. POU5F1/Oct-4 expression in breast cancer tissue is significantly associated with non-sentinel lymph node metastasis. BMC Cancer 2016, 16, 175. [Google Scholar] [CrossRef] [PubMed]
- Hicklin, D.J.; Marincola, F.M.; Ferrone, S. HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story. Mol. Med. Today 1999, 5, 178–186. [Google Scholar] [CrossRef]
- Di Sabatino, A.; Corazza, G.R. Celiac disease. Lancet 2009, 373, 1480–1493. [Google Scholar] [CrossRef]
- CherÑavsky, A.C.; Rubio, A.E.; Vanzulli, S.; Rubinstein, N.; Rosa, S.D.; Fainboim, L. Evidences of the involvement of Bak, a member of the Bcl-2 family of proteins, in active coeliac disease. Autoimmunity 2002, 35, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Spies, T.; Bresnahan, M.; Strominger, J.L. Human major histocompatibility complex contains a minimum of 19 genes between the complement cluster and HLA-B. Proc. Natl. Acad. Sci. USA 1989, 86, 8955–8958. [Google Scholar] [CrossRef] [PubMed]
- Maranto, A.R. Primary structure, ligand binding, and localization of the human type 3 inositol 1,4,5-trisphosphate receptor expressed in intestinal epithelium. J. Biol. Chem. 1994, 269, 1222–1230. [Google Scholar] [PubMed]
- Sumitani, M.; Kasashima, K.; Ohta, E.; Kang, D.; Endo, H. Association of a novel mitochondrial protein M19 with mitochondrial nucleoids. J. Biochem. 2009, 146, 725–732. [Google Scholar] [CrossRef] [PubMed]
| Combination | SNP 1 | SNP 2 | ... | SNP k | Case | Control | Total |
|---|---|---|---|---|---|---|---|
| 1 | AA | BB | KK | ||||
| 2 | Aa | BB | KK | ||||
| 3 | aa | BB | KK | ||||
| : | : | : | : | : | : | : | : |
| : | : | : | : | : | : | : | : |
| aa | bb | Kk | : | : | : | ||
| aa | bb | kk | |||||
| Total | N |
| Significant Combination | Chromosome and Related Genes | Single-Locus p-Value | Combination p-Value |
|---|---|---|---|
| (rs1108842, rs4687657) | (chr3: GNL3, chr3: ITIH4) | (, ) | |
| (rs4408545, rs3785181) | (chr16: AFG3L1P, chr16: GAS11) | (, ) | |
| (rs3811040, rs2723192) | (chr2: CKAP2L, chr2: IL37) | (, ) | |
| (rs9379968, rs204994) | (chr6: *, chr6: AGER) | (, ) | |
| (rs9257694, rs3129943) | (chr6: OR14J1, chr6: LOC101929163) | (, ) | |
| (rs879882, rs2523608, rs592229) | (chr6:POU5F1, chr6:HLA-B, chr6:SKIV2L) | (, , ) |
| Significant Combination | Chromosome and Related Genes | Single-Locus p-Value | Combination p-Value |
|---|---|---|---|
| (rs2844509, rs9262495) | (chr6: DDX39B, chr6: MUC22) | , ) | |
| (rs2256028, rs406936) | (chr6: MICA, chr6: SKIV2L) | (, ) | |
| (rs210138, rs2894342) | (chr6: BAK1, Chr6:*) | (, ) | |
| (rs3130785, rs2844509) | (chr6: LINC00243, chr6: DDX39B) | (, ) | |
| (rs1519643, rs1481417) | (chr2: *, chr14:*) | (, ) | |
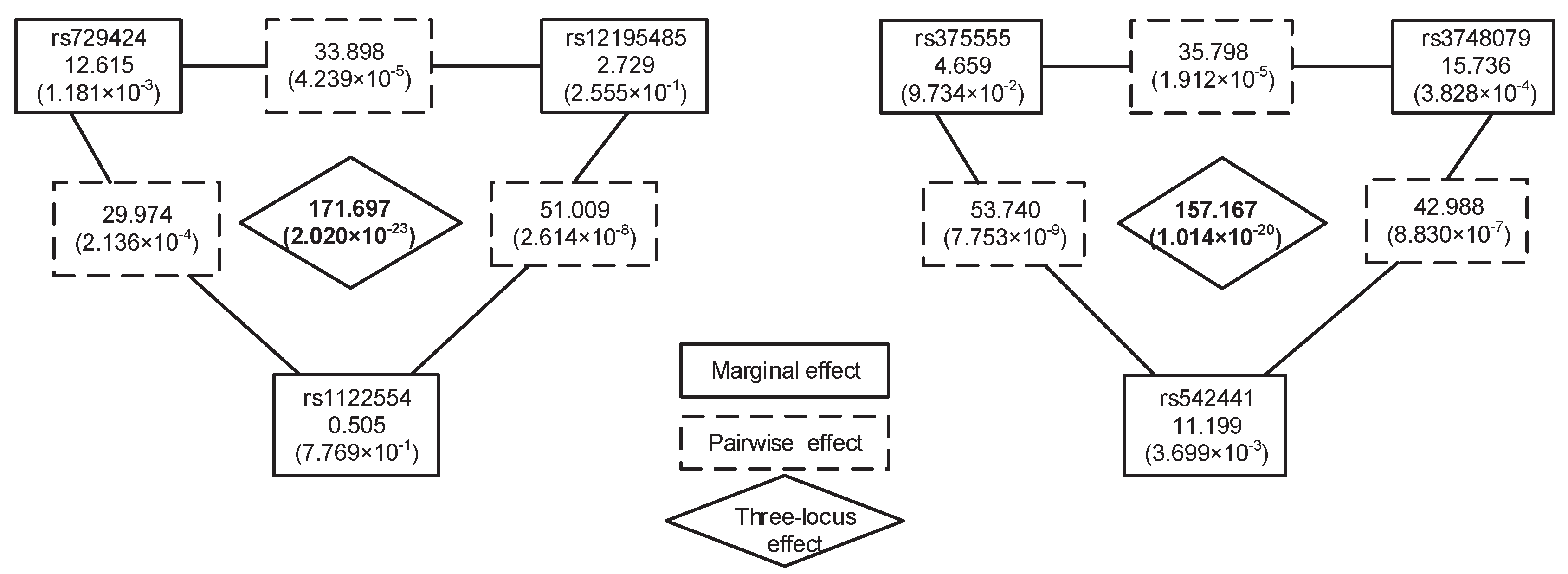
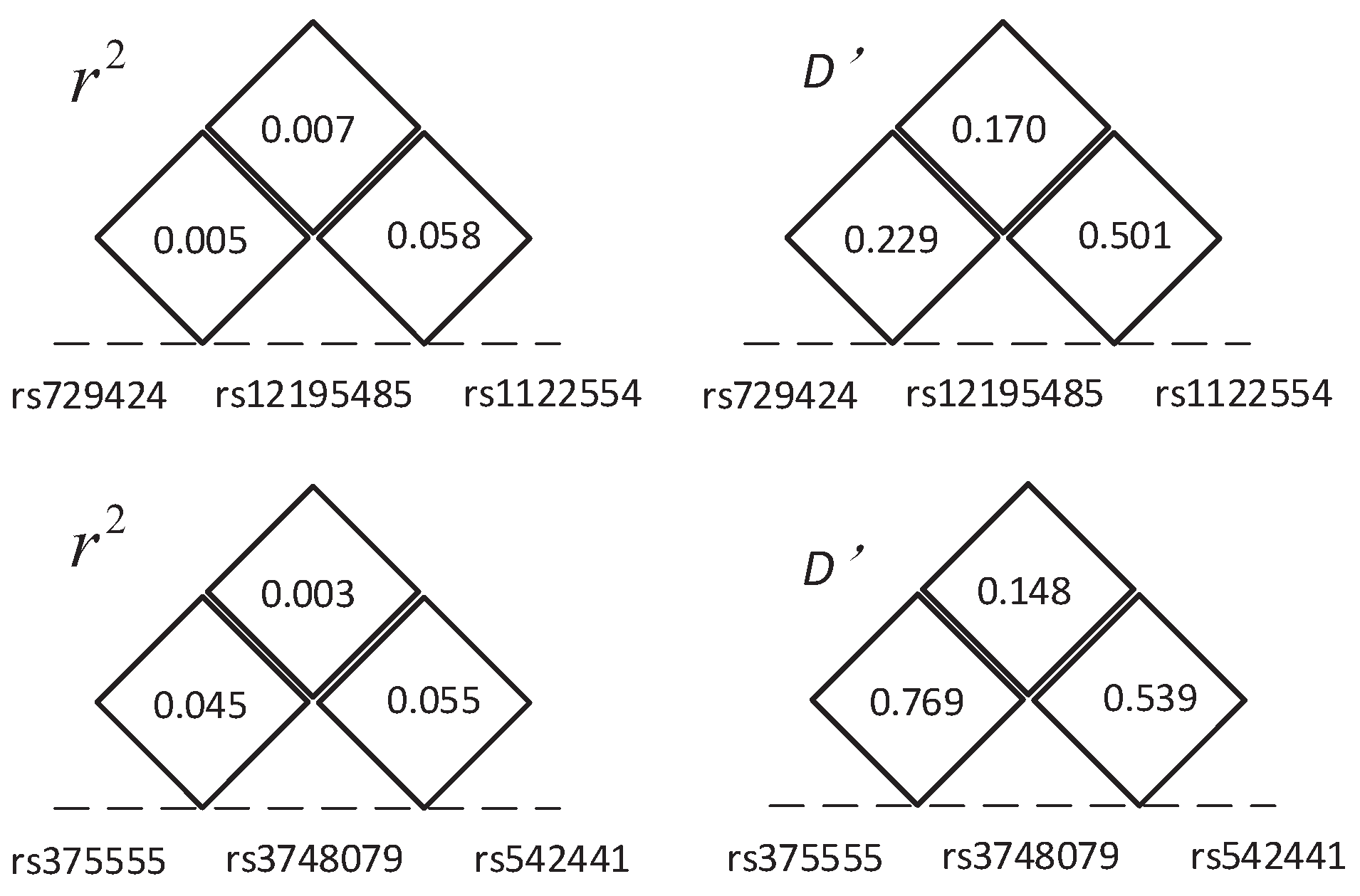
| (rs729424, rs12195485, rs1122554) | (chr6:ITPR3, chr6:LOC105375025, chr:*) | (, , ) | |
| (rs375555, rs3748079, rs542441) | (chr6: *, chr6: ITPR3, chr6: UQCC2) | (, , ) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Yu, G.; Jiang, Y.; Wang, J. HiSeeker: Detecting High-Order SNP Interactions Based on Pairwise SNP Combinations. Genes 2017, 8, 153. https://doi.org/10.3390/genes8060153
Liu J, Yu G, Jiang Y, Wang J. HiSeeker: Detecting High-Order SNP Interactions Based on Pairwise SNP Combinations. Genes. 2017; 8(6):153. https://doi.org/10.3390/genes8060153
Chicago/Turabian StyleLiu, Jie, Guoxian Yu, Yuan Jiang, and Jun Wang. 2017. "HiSeeker: Detecting High-Order SNP Interactions Based on Pairwise SNP Combinations" Genes 8, no. 6: 153. https://doi.org/10.3390/genes8060153





