Genome-Wide Comprehensive Analysis the Molecular Phylogenetic Evaluation and Tissue-Specific Expression of SABATH Gene Family in Salvia miltiorrhiza
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of the Members of SABATH Gene Family in S. miltiorrhiza
2.2. Sequence Feature and Gene Structure Analyses
2.3. Multiple Sequence Alignment, Phylogenetic Analyses and Motif Detection
2.4. Ka and Ks Calculation
2.5. Tests of Positive Selection
2.6. Estimation of Functional Divergence
2.7. Plant Materials, RNA Extraction and Real-Time qPCR
3. Results and Discussion
3.1. Sequence Feature of SABATH Genes in S. miltiorrhiza
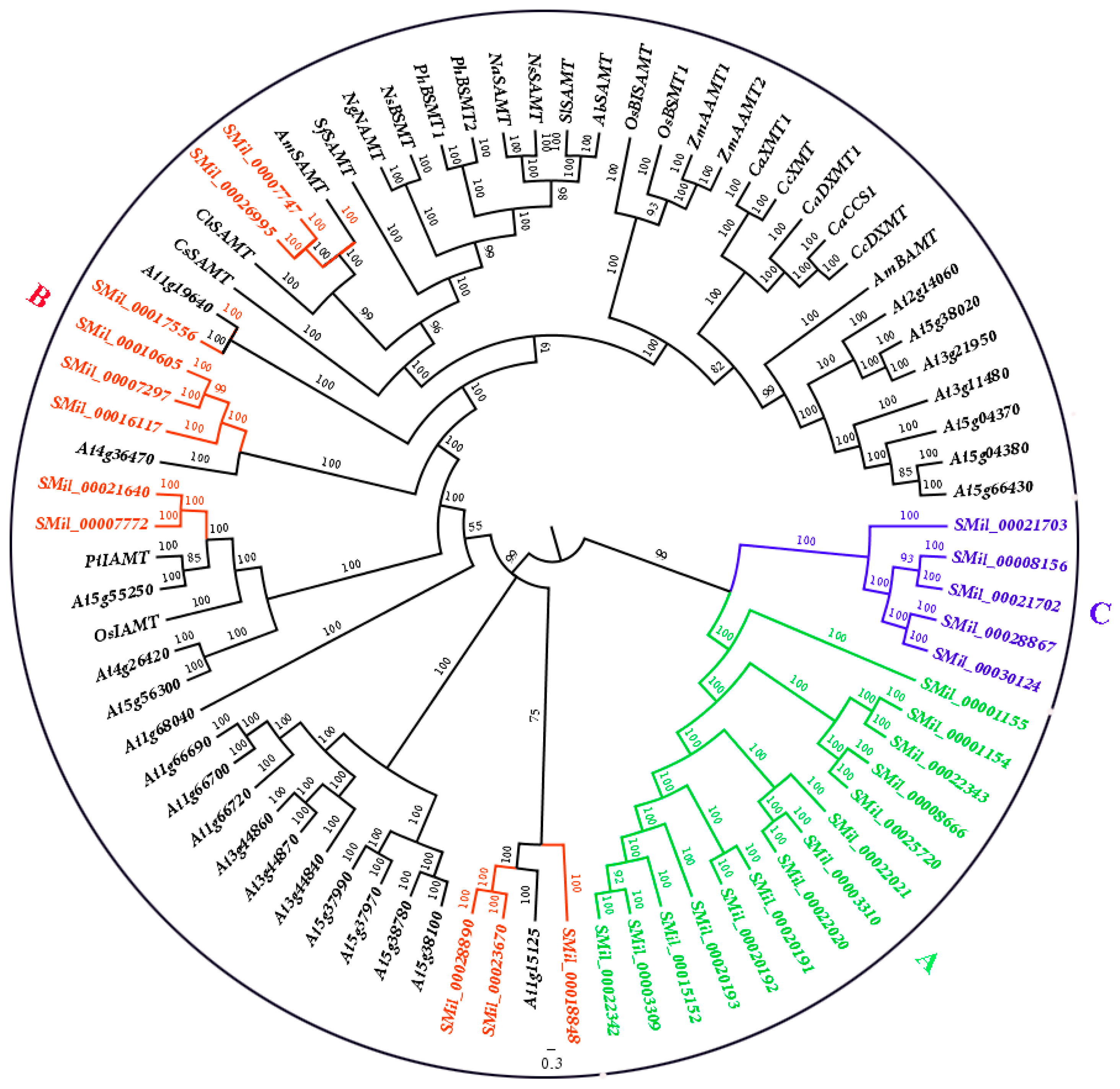
3.2. Phylogenetic Analysis of SABATH Gene Family in S. miltiorrhiza
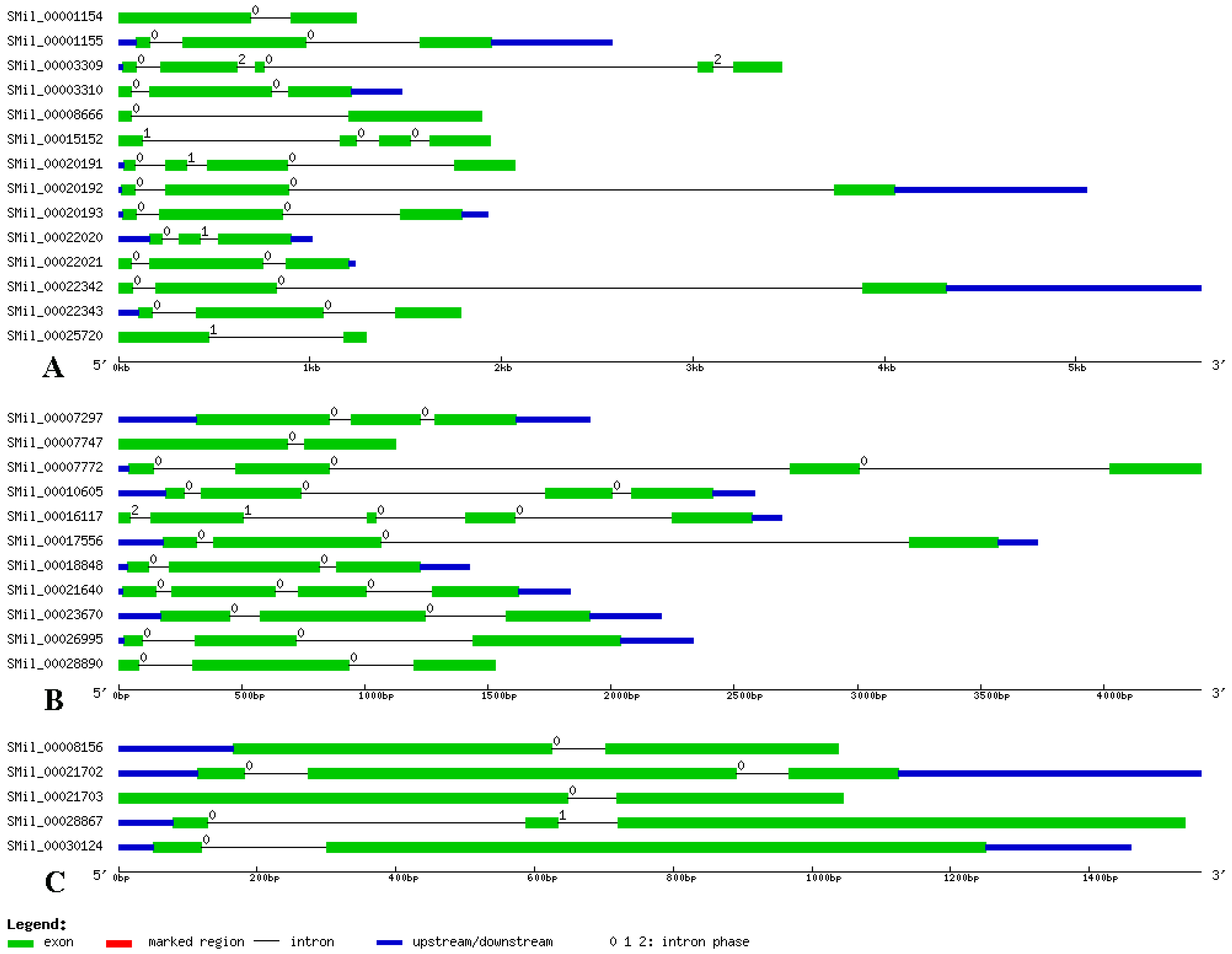
3.3. Gene Structure Analysis of SABATH Genes in S. miltiorrhiza
3.4. Analysis of Conserved Domains and Motifs
3.5. Driving Forces for Genetic Divergence
3.6. Positive Selection on SmSABATH Genes
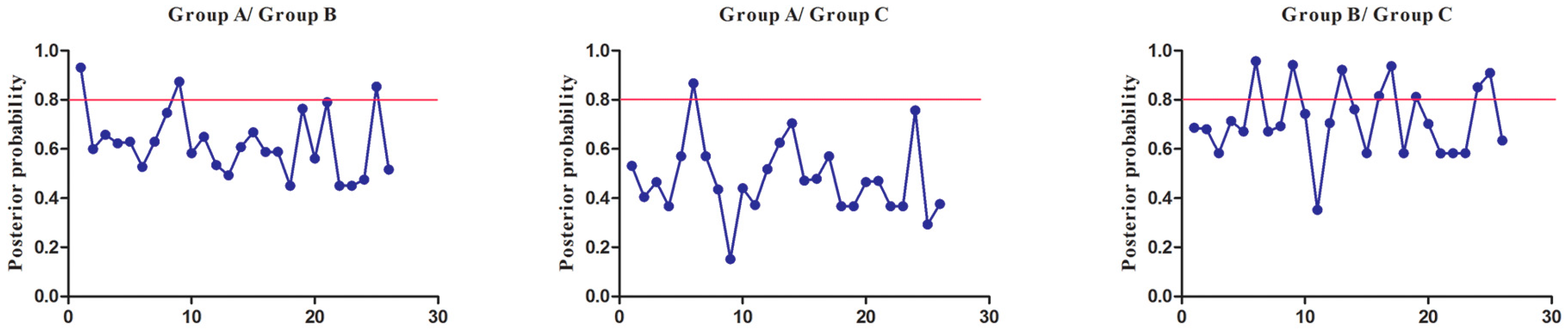
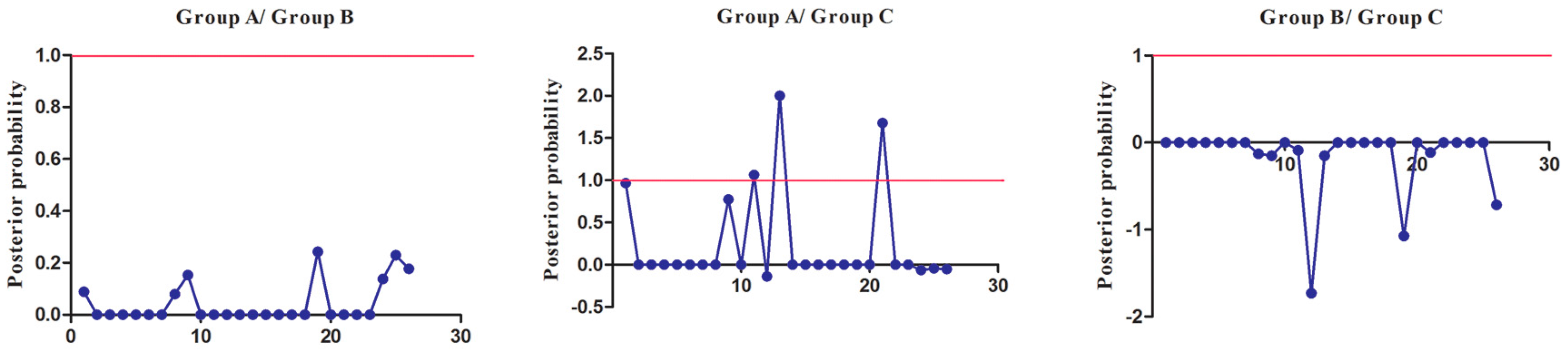
3.7. Functional Divergence Analysis (FDA) of SmSABATH Proteins
3.8. Tissue-Specific Expression of SABATH Gene Family in S. miltiorrhiza
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Attieh, J.; Djiana, R.; Koonjul, P.; Étienne, C.; Sparace, S.A.; Saini, H.S. Cloning and functional expression of two plant thiol methyltransferases: A new class of enzymes involved in the biosynthesis of sulfur volatiles. Plant Mol. Biol. 2002, 50, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Effmert, U.; Saschenbrecker, S.; Ross, J.; Negre, F.; Fraser, C.M.; Noel, J.P.; Dudareva, N.; Piechulla, B. Floral benzenoid carboxyl methyltransferases: From in vitro to in planta function. Phytochemistry 2005, 66, 1211–1230. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, C. Structural basis for the modulation of lignin monomer methylation by caffeic Acid/5-Hydroxyferulic Acid 3/5-O-Methyltransferase. Plant Cell Online 2002, 14, 1265–1277. [Google Scholar] [CrossRef]
- Zubieta, C. Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell Online 2003, 15, 1704–1716. [Google Scholar] [CrossRef]
- Zubieta, C.; He, X.Z.; Dixon, R.A.; Noel, J.P. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 2001, 8, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Noel, J.P.; Dixon, R.A.; Pichersky, E.; Zubieta, C.; Ferrer, J.L. Chapter two Structural, functional, and evolutionary basis for methylation of plant small molecules. Recent Adv. Phytochem. 2003, 37, 37–58. [Google Scholar]
- Ross, J.R.; Nam, K.H.; D’Auria, J.C.; Pichersky, E. S-adenosyl-l-methionine: Salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 1999, 367, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Murfitt, L.M.; Kolosova, N.; Mann, C.J.; Dudareva, N. Purification and characterization of S-adenosyl-l-methionine: Benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methyl benzoate in flowers of Antirrhinum majus. Arch. Biochem. Biophys. 2000, 382, 145–151. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, J.C.; Chen, F.; Pichersky, E. Chapter eleven the SABATH family of MTS in Arabidopsis thaliana and other plant species. Recent Adv. Phytochem. 2003, 37, 253–283. [Google Scholar]
- Zhao, N.; Ferrer, J.L.; Moon, H.S.; Kapteyn, J.; Zhuang, X.; Hasebe, M.; Stewart, C.N., Jr.; Gang, D.R.; Chen, F. A SABATH Methyltransferase from the moss Physcomitrella patens catalyzes S-methylation of thiols and has a role in detoxification. Phytochemistry 2012, 81, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.S.; Song, J.T.; Cheong, J.J.; Lee, Y.H.; Lee, Y.W.; Hwang, I.; Lee, J.S.; Choi, Y.D. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Zhao, Y.; Ma, Z.; Shi, G.; Yang, Y.; Pichersky, E.; Chen, H.; Liu, M.; Chen, Z.; et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 2005, 17, 2693–2704. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yuan, J.S.; Ross, J.; Noel, J.P.; Pichersky, E.; Chen, F. An Arabidopsis thaliana methyltransferase capable of methylating farnesoic acid. Arch. Biochem. Biophys. 2006, 448, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Varbanova, M.; Yamaguchi, S.; Yang, Y.; McKelvey, K.; Hanada, A.; Borochov, R.; Yu, F.; Jikumaru, Y.; Ross, J.; Cortes, D.; et al. Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 2007, 19, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.; Qualley, A.V.; Xie, Z.; Fridman, E.; Dudareva, N.; Gang, D.R. Evolution of cinnamate/p-coumarate carboxyl methyltransferases and their role in the biosynthesis of methylcinnamate. Plant Cell 2007, 19, 3212–3229. [Google Scholar] [CrossRef] [PubMed]
- Murata, J.; Roepke, J.; Gordon, H.; De Luca, V. The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 2008, 20, 524–542. [Google Scholar] [CrossRef] [PubMed]
- Kollner, T.G.; Lenk, C.; Zhao, N.; Seidl-Adams, I.; Gershenzon, J.; Chen, F.; Degenhardt, J. Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using S-adenosyl-l-methionine. Plant Physiol. 2010, 153, 1795–1807. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Herai, Y.; Koizumi, N.; Kusano, T.; Sano, H. 7-Methylxanthine methyltransferase of coffee plants. Gene isolation and enzymatic properties. J. Biol. Chem. 2000, 276, 8213–8218. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Boyle, B.; Duval, I.; Ferrer, J.L.; Lin, H.; Seguin, A.; MacKay, J.; Chen, F. SABATH methyltransferases from white spruce (Picea glauca): Gene cloning, functional characterization and structural analysis. Tree Physiol. 2009, 29, 947–957. [Google Scholar] [CrossRef] [PubMed]
- And, R.A.C.; Mullet, J.E. Biosynthesis and action of jasmonates in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 355–381. [Google Scholar]
- Wasternack, C.; Hause, B. Jasmonates and octadecanoids: Signals in plant stress responses and development. Prog. Nucleic Acid Res. Mol. Biol. 2002, 72, 165–221. [Google Scholar] [PubMed]
- Ljung, K.; Hull, A.K.; Kowalczyk, M.; Marchant, A.; Celenza, J.; Cohen, J.D.; Sandberg, G. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. In Auxin Molecular Biology; Springer: Dordrecht, The Netherlands, 2002; pp. 249–272. [Google Scholar]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; D’Auria, J.C.; Tholl, D.; Ross, J.R.; Gershenzon, J.; Noel, J.P.; Pichersky, E. An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. Cell Mol. Biol. 2003, 36, 577–588. [Google Scholar] [CrossRef]
- Xu, R.; Song, F.; Zheng, Z. OsBISAMT1, a gene encoding S-adenosyl-l-methionine: Salicylic acid carboxyl methyltransferase, is differentially expressed in rice defense responses. Mol. Biol. Rep. 2006, 33, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Uefuji, H.; Tatsumi, Y.; Morimoto, M.; Kaothien-Nakayama, P.; Ogita, S.; Sano, H. Caffeine production in tobacco plants by simultaneous expression of three coffee N-methyltrasferases and its potential as a pest repellant. Plant Mol. Biol. 2005, 59, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Seifert, K.; Unger, W. Insecticidal and fungicidal compounds from Isatis tinctoria. Z. Naturforsch. C J. Biosci. 1994, 49, 44. [Google Scholar] [CrossRef]
- Li, Y.G.; Song, L.; Liu, M.; Hu, Z.B.; Wang, Z.T. Advancement in analysis of Salviae miltiorrhizae Radix et Rhizoma (Danshen). J. Chromatogr. A 2009, 1216, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yuan, L.; Wu, B.; Li, X.; Chen, S.; Lu, S. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 2012, 63, 2809–2823. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.H.; Chen, A.H.; Zhang, B.L. Salviae Miltiorrhiza: A model organism for chinese traditional medicine genomic studies. Acta Chin. Med. Pharmacol. 2009, 37, 1–4. [Google Scholar]
- Xu, H.; Song, J.; Luo, H.; Zhang, Y.; Li, Q.; Zhu, Y.; Xu, J.; Li, Y.; Song, C.; Wang, B. Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. Plant 2016, 9, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Luo, H.M.; Li, C.F.; Sun, C.; Xu, J.; Chen, S.L. Salvia miltiorrhiza as medicinal model plant. Acta Pharm. Sin. 2013, 48, 1099–1106. [Google Scholar]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffithsjones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2004, 32, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G. Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Mol. Biol. Evol. 2005, 22, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Tchieu, J.; Saier, S.M., Jr. A web-based Tree View (TV) program for the visualization of phylogenetic trees. J. Mol. Microbiol. Biotechnol. 2002, 4, 69–70. [Google Scholar] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, H.; Xu, Z.; Zhu, Y.; Ji, A.; Song, J.; Chen, S. Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci. Rep. 2015, 5, 11244. [Google Scholar] [CrossRef] [PubMed]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef] [PubMed]
- Fares, M.A. SWAPSC: Sliding window analysis procedure to detect selective constraints. Bioinformatics 2004, 20, 2867–2868. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R.; Goldman, N.; Pedersen, A.M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 2000, 155, 431–449. [Google Scholar] [PubMed]
- Zhang, J.; Nielsen, R.; Yang, Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005, 22, 2472–2479. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zou, Y.; Su, Z.; Huang, W.; Zhou, Z.; Arendsee, Z.; Zeng, Y. An update of DIVERGE software for functional divergence analysis of protein family. Mol. Biol. Evol. 2013, 30, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, D.; Shao, F.; Lu, S. Molecular cloning and expression analysis of WRKY transcription factor genes in Salvia miltiorrhiza. BMC Genom. 2015, 16, 200. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 44, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Gu, X. Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 1999, 16, 1664–1674. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative Ct method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hou, S.; Cui, G.; Chen, S.; Wei, J.; Huang, L. Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol. Biol. Rep. 2010, 37, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, Y.; Wang, X.; Gu, S.; Yu, J.; Liang, G.; Yan, C.; Xu, C. Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics 2008, 92, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. The exon theory of genes. Cold Spring Harb. Symp. Quant. Biol. 1987, 52, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Patthy, L. Intron-dependent evolution: Preferred types of exons and introns. FEBS Lett. 1987, 214, 1–7. [Google Scholar] [CrossRef]
- Joshi, C.P.; Chiang, V.L. Conserved sequence motifs in plant S-adenosyl-l-methionine-dependent methyltransferases. Plant Mol. Biol. 1998, 37, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Kagan, R.M.; Clarke, S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 1994, 310, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zheng, H.; Yang, S.; Yu, H.; Li, J.; Jiang, H.; Su, J.; Yang, L.; Zhang, J.; Mcdermott, J. Origin and evolution of new exons in rodents. Genome Res. 2005, 15, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Gu, X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol. Biol. Evol. 2006, 23, 1937–1945. [Google Scholar] [CrossRef] [PubMed]



| Model | np | lnL | Estimates of Parameter 1 | 2ΔlnL | Positive Selection Sites 2 | |
|---|---|---|---|---|---|---|
| Frequency | dN/dS | |||||
| M0 (one-ratio) | 60 | −24,100.940606 | 0.32227 | None | ||
| M3 (discrete) | 64 | −23,631.868500 | p0 = 0.15469 p1 = 0.41191 p2 = 0.43340 | ω0 = 0.06316 ω1 = 0.25310 ω2 = 0.65803 | 938.1442 (M3 vs. M0) ** | Not allowed |
| M1a (nearly neutral) | 61 | −23,766.855086 | p0 = 0.53255 p1 = 0.46745 | ω0 = 0.23043 ω1 = 1.00000 | None | |
| M2a (positive selection) | 63 | −23,766.855086 | p0 = 0.53255 p1 = 0.30714 p2 = 0.16031 | ω0 = 0.23043 ω1 = 1.00000 ω2 = 1.00000 | 0 (M2a vs. M1a) | Not allowed |
| M7 (beta) | 61 | −23,622.773935 | p = 1.10441 q = 1.67064 | None | ||
| M8 (beta & ω) | 63 | −23,622.775486 | p0 = 0.99999 p = 1.10448 q = 1.67080 p1 = 0.00001 | ω = 5.70606 | 0.00308 (M8 vs. M7) | 181 Q 432 V 434 G 460 S 476 H 479 C |
| Foreground Branches | Branch-Site Model | lnL | 2ΔlnL | p Value | ω Values 1 | Positively Selected Sites 2 |
|---|---|---|---|---|---|---|
| Group A | Null | −23,766.855109 | 0 | 1 | ω0 = 0.23043 ω1 = 1.00000 ω2 = 1.00000 | None |
| Alternative | −23,766.855151 | ω0 = 0.23043 ω1 = 1.00000 ω2 = 3.05353 | ||||
| Group B | Null | −23,766.855088 | 5.653506 | 0.017 | ω0 = 0.23043 ω1 = 1.00000 ω2 = 1.00000 | 152 A 0.649 209 A 0.815 385 R 0.516 |
| Alternative | −23,764.028335 | ω0 = 0.23051 ω1 = 1.00000 ω2 = 464.10117 | ||||
| Group C | Null | −23,766.466691 | 12.07159 | <0.01 | ω0 = 0.22946 ω1 = 1.00000 ω2 = 1.00000 | 137 K 0.772 200 I 0.514 252 H 0.889 461 A 0.961 * |
| Alternative | −23,760.430894 | ω0 = 0.22951 ω1 = 1.00000 ω2 = 998.97795 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Wang, S.; Wang, Z. Genome-Wide Comprehensive Analysis the Molecular Phylogenetic Evaluation and Tissue-Specific Expression of SABATH Gene Family in Salvia miltiorrhiza. Genes 2017, 8, 365. https://doi.org/10.3390/genes8120365
Wang B, Wang S, Wang Z. Genome-Wide Comprehensive Analysis the Molecular Phylogenetic Evaluation and Tissue-Specific Expression of SABATH Gene Family in Salvia miltiorrhiza. Genes. 2017; 8(12):365. https://doi.org/10.3390/genes8120365
Chicago/Turabian StyleWang, Bin, Shiqiang Wang, and Zhezhi Wang. 2017. "Genome-Wide Comprehensive Analysis the Molecular Phylogenetic Evaluation and Tissue-Specific Expression of SABATH Gene Family in Salvia miltiorrhiza" Genes 8, no. 12: 365. https://doi.org/10.3390/genes8120365




