Assessment of Bifidobacterium Species Using groEL Gene on the Basis of Illumina MiSeq High-Throughput Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Media and DNA Extraction
2.2. Fecal Sample Collection and Genomic DNA Extraction
2.3. Phylogenetic Analysis
2.4. Bifidobacterium groEL-Specific Primer Design
2.5. PCR Amplification, Quantification, and Sequencing
2.6. Evaluation of the Sensitivity of the Novel Designed Primer Set
2.7. Bioinformatic Sequence Analysis
2.8. Real-Time qPCR
2.9 Statistical Analysis
3. Results
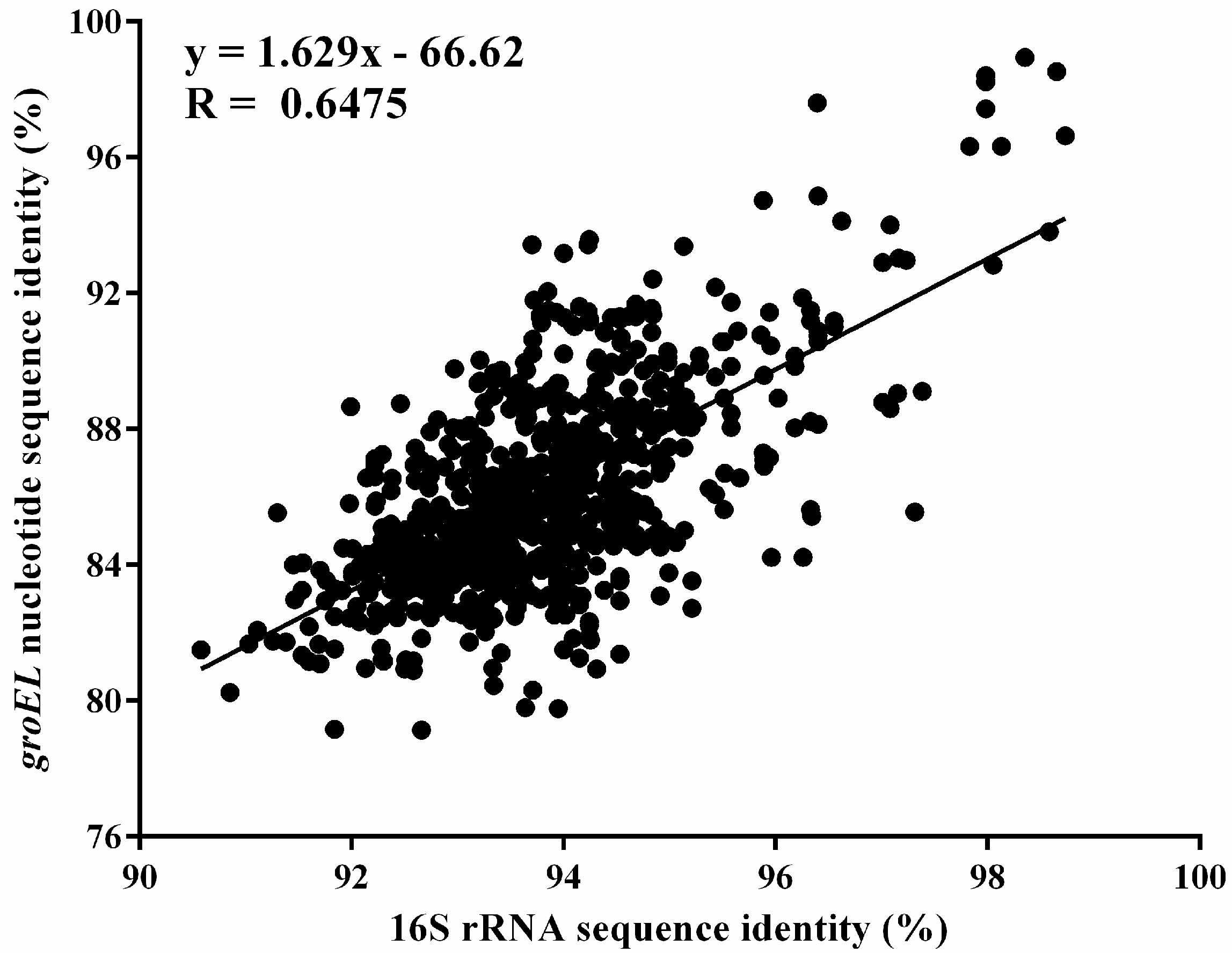
3.1. Comparative Analysis of the groEL and 16S rRNA Gene
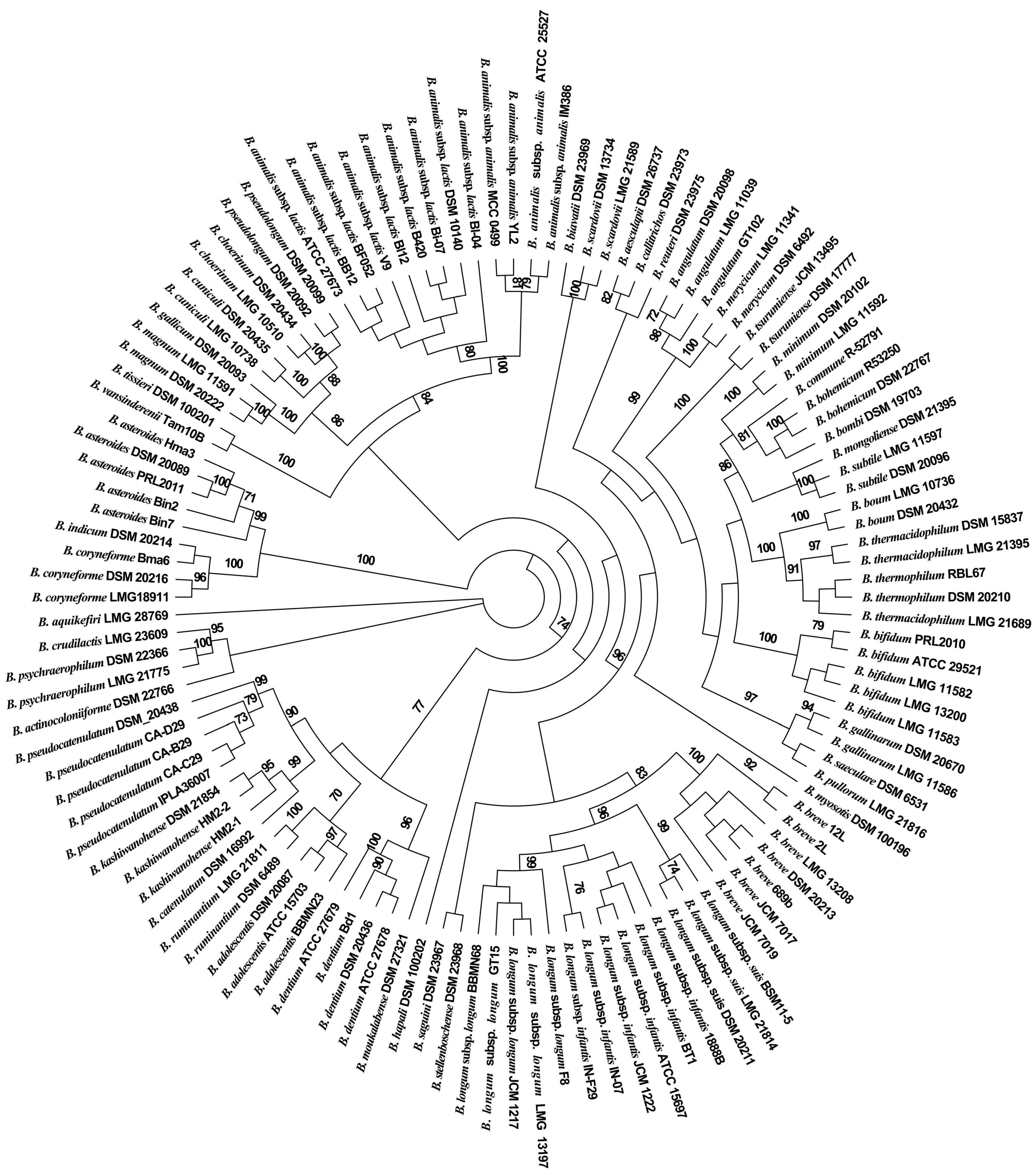
3.2. Phylogenetic Analysis of the Partial groEL Gene
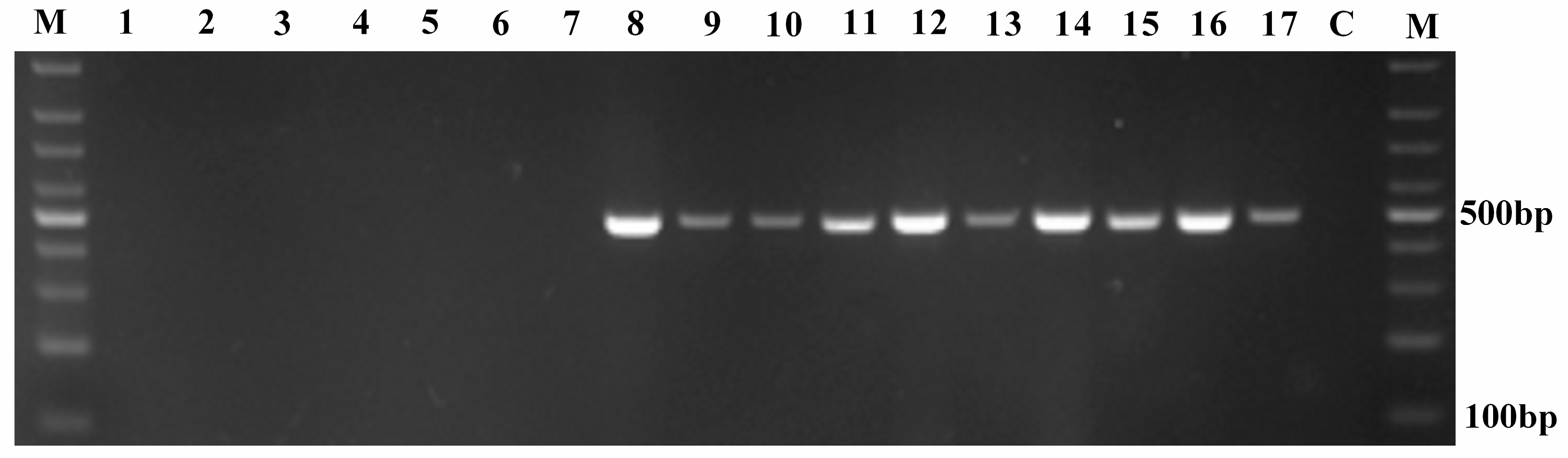
3.3. Specificity, Accuracy and Sensitivity of the Novel Designed Primer Set
3.4. Comparison of Resolving Power between the Partial groEL Gene and V3–V4 Region of 16S rRNA
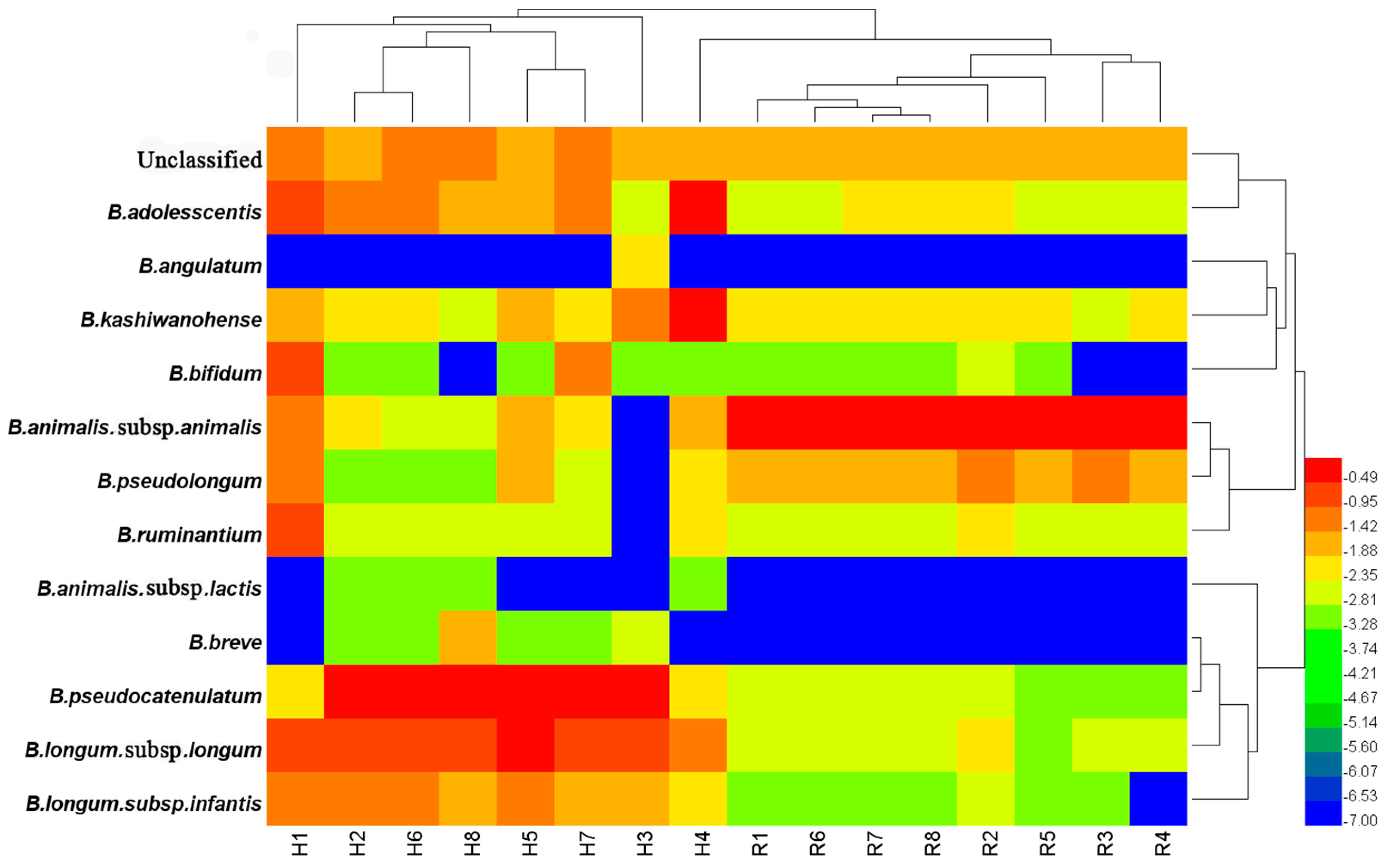
3.5. Comparison of Bifidobacterium Species between Humans and Rats
3.6. qPCR-Based Determination of Main Bifidobacterial Numbers
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Arvanitoyannis, I.S.; Van Houwelingen-Koukaliaroglou, M. Functional foods: A survey of health claims, pros and cons, and current legislation. Crit. Rev. Food Sci. Nutr. 2005, 45, 385–404. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Marchesi, J.R.; Foroni, E.; Gueimonde, M.; Shanahan, F.; Margolles, A.; van Sinderen, D.; Ventura, M. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 2009, 3, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C.; et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Hevia, A.; Foroni, E.; Duranti, S.; Turroni, F.; Lugli, G.A.; Sanchez, B.; Martín, R.; Gueimonde, M.; van Sinderen, D.; et al. Assessing the fecal microbiota: An optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS ONE 2013, 8, e68739. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Li, D.; Ai, C.; Zhao, J.; Zhang, H.; Chen, W. Lactulose differently modulates the composition of luminal and mucosal microbiota in C57BL/6J mice. J. Agric. Food Chem. 2016, 64, 6240–6247. [Google Scholar] [CrossRef] [PubMed]
- Loman, N.J.; Misra, R.V.; Dallman, T.J.; Constantinidou, C.; Gharbia, S.E.; Wain, J.; Pallen, M.J. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012, 30, 434–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milani, C.; Lugli, G.A.; Turroni, F.; Mancabelli, L.; Duranti, S.; Viappiani, A.; Mangifesta, M.; Segata, N.; van Sinderen, D.; Ventura, M. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol. Ecol. 2014, 90, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Stewart, F.J.; Cavanaugh, C.M. Intragenomic variation and evolution of the internal transcribed spacer of the rRNA operon in bacteria. J. Mol. Evol. 2007, 65, 44–67. [Google Scholar] [CrossRef] [PubMed]
- Crosby, L.D.; Criddle, C.S. Understanding bias in microbial community analysis techniques due to RRN operon copy number heterogeneity. Biotechniques 2003, 34, 790–802. [Google Scholar] [PubMed]
- Ventura, M.; Canchaya, C.; van Sinderen, D.; Fitzgerald, G.F.; Zink, R. Bifidobacterium lactis DSM 10140: Identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 2004, 70, 3110–3121. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Zink, R. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 2003, 69, 7517–7522. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Meylan, V.; Klaenhammer, T.R.; Zink, R. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 2003, 69, 6908–6922. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Zink, R.; Fitzgerald, G.F.; van Sinderen, D. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl. Environ. Microbiol. 2005, 71, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Requena, T.; Burton, J.; Matsuki, T.; Munro, K.; Simon, M.A.; Tanaka, R.; Watanabe, K.; Tannock, G.W. Identification, detection, and enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 2002, 68, 2420–2427. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Chambers, J.R.; Barlow, K.; Park, A.S.; Wheatcroft, R. The gene encoding xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (xfp) is conserved among Bifidobacterium species within a more variable region of the genome and both are useful for strain identification. FEMS Microbiol. Lett. 2005, 246, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Del Casale, A.; Dellaglio, F.; Neviani, E.; Fitzgerald, G.F.; van Sinderen, D. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 2006, 30, 734–759. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, W.; Dong, X. Species identification of genus Bifidobacterium based on partial HSP60 gene sequences and proposal of Bifidobacterium thermacidophilum subsp. porcinum subsp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 1619–1623. [Google Scholar] [CrossRef] [PubMed]
- Masco, L.; Ventura, M.; Zink, R.; Huys, G.; Swings, J. Polyphasic taxonomic analysis of Bifidobacterium animalis and Bifidobacterium lactis reveals relatedness at the subspecies level: Reclassification of Bifidobacterium animalis as Bifidobacterium animalis subsp. animalis subsp. nov. and Bifidobacterium lactis as Bifidobacterium animalis subsp. lactis subsp. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1137–1143. [Google Scholar] [PubMed]
- Ventura, M.; Canchaya, C.; Zink, R.; Fitzgerald, G.F.; van Sinderen, D. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: Genetic, transcriptional, and phylogenetic analyses. Appl. Environ. Microbiol. 2004, 70, 6197–6209. [Google Scholar] [CrossRef] [PubMed]
- Junick, J.; Blaut, M. Quantification of human fecal Bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl. Environ. Microbiol. 2012, 78, 2613–2622. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.E.; Penny, S.L.; Crowell, K.G.; Goh, S.H.; Hemmingsen, S.M. cpnDB: A chaperonin sequence database. Genome Res. 2004, 14, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Schell, M.A.; Karmirantzou, M.; Snel, B.; Vilanova, D.; Berger, B.; Pessi, G.; Zwahlen, M.C.; Desiere, F.; Bork, P.; Delley, M.; et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 2002, 99, 14422–14427. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, C.; Johansen, E.; Pedersen, M.B. Complete genome sequence of Bifidobacterium animalis subsp. lactis BB-12, a widely consumed probiotic strain. J. Bacteriol. 2010, 192, 2467–2468. [Google Scholar] [CrossRef] [PubMed]
- Zhurina, D.; Zomer, A.; Gleinser, M.; Brancaccio, V.F.; Auchter, M.; Waidmann, M.S.; Westermann, C.; van Sinderen, D.; Riedel, C.U. Complete genome sequence of Bifidobacterium bifidum S17. J. Bacteriol. 2011, 193, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Reniero, R.; Zink, R. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 2001, 67, 2760–2765. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.R.; Geng, L.L.; Li, Y.H.; Wang, Q.; Diao, Q.Y.; Zhou, T.; Dai, P.L. The effects of Bt Cry1Ie toxin on bacterial diversity in the midgut of Apis mellifera ligustica (Hymenoptera: Apidae). Sci. Rep. 2016, 6, 24664. [Google Scholar] [CrossRef] [PubMed]
- Staroscik, A. Calculator for Determining the Number of Copies of a Template. Available online: http://cels.uri.edu/gsc/cndna.html (accessed on 8 February 2017).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011. [Google Scholar]
- Lewisa, Z.T.; Bokulicha, N.A.; Kalanetraa, K.M.; RuizMoyanoa, S.; Underwoodb, M.A.; Mills, D.A. Use of bifidobacterial specific terminal restriction fragment length polymorphisms to complement next generation sequence profiling of infant gut communities. Anaerobe 2013, 19, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Miyake, T.; Watanabe, K.; Watanabe, T.; Oyaizu, H. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol. Immunol. 1998, 42, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Jian, W.; Zhu, L.; Dong, X. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 2001, 51, 1633–1638. [Google Scholar] [CrossRef] [PubMed]
- Duranti, S.; Mangifesta, M.; Lugli, G.A.; Turroni, F.; Anzalone, R.; Milani, C.; Mancabelli, L.; Ossiprandi, M.C.; Ventura, M. Bifidobacterium vansinderenii sp. nov., isolated from faeces of emperor tamarin (Saguinus imperator). Int. J. Syst. Evol. Microbiol. 2017, 67, 3987–3995. [Google Scholar] [CrossRef] [PubMed]



| Number | Bifidobacterium Species | Strain a | GenBank Accession no. of groEL Gene Sequences | GenBank Accession no. of 16S rRNA Gene Sequences |
|---|---|---|---|---|
| 1 | B. adolescentis | ATCC 15703 | AP009256 | NR_074802 |
| 2 | B. angulatum | JCM 7096 | AP012322 | LC071846 |
| 3 | B. animalis subsp. animalis | ATCC 25527 = LMG 10508 | CP002567 | JGYM01000004 |
| 4 | B. animalis subsp. lactis | BB12 | CP001853 | GU116483 |
| 5 | B. asteroides | Bin2 | NZ_KQ033859 | EF187231 |
| 6 | B. biavatii | DSM 23969 | JGYN01000004 | JGYN01000007 |
| 7 | B. bifidum | PRL2010 | CP001840 | CP001840 |
| 8 | B. bohemicum | R53250 | FMAM01000001 | FMAM01000014 |
| 9 | B. boum | LMG 10736 = JCM 1211 | JGYQ01000016 | LC071799 |
| 10 | B. breve | JCM 1192 | AP012324 | LC071793 |
| 11 | B. callitrichos | DSM 23973 | JGYS01000001 | JGYS01000004 |
| 12 | B. catenulatum | DSM 16992 | AP012325 | NR_041875 |
| 13 | B. choerinum | ATCC 27686 = LMG 10510 | JGYU01000001 | D86186 |
| 14 | B. coryneforme | Bma6 | KQ033865 | EF187237 |
| 15 | B. crudilactis | LMG 23609 | NZ_JHAL01000002 | NZ_JHAL01000001 |
| 16 | B. cuniculi | LMG 10738 | JGYV01000008 | JX986964 |
| 17 | B. dentium | JCM 1195 | AP012326 | LC071795 |
| 18 | B. gallicum | DSM 20093 = LMG 11596 | NZ_ABXB03000002 | ABXB03000004 |
| 19 | B. gallinarum | DSM 20670 = JCM 6291 | NZ_JDUN01000004 | D86191 |
| 20 | B. indicum | LMG 11587 = DSM 20214 = JCM1302 | CP006018 | D86188 |
| 21 | B. magnum | DSM 20222 = JCM 1218 | NZ_ATVE01000001 | D86193 |
| 22 | B. merycicum | DSM 6492 = JCM 8219 | NZ_JDTL01000006 | D86192 |
| 23 | B. minimum | DSM 20102 = ATCC 27538 | NZ_ATXM01000001 | M58741 |
| 24 | B. kashiwanohense | HM2-1 | AB578933 | AB491757 |
| 25 | B. longum subsp. infantis | ATCC 15697 | CP001095 | NR_043437 |
| 26 | B. longum subsp. longum | BBMN68 | CP002286 | GQ380695.1 |
| 27 | B. mongoliense | DSM 21395 | JGZE01000001 | AB433856 |
| 28 | B. pseudocatenulatum | JCM 1200 | AP012330 | LC071796 |
| 29 | B. pseudolongum | PV8-2 | CP007457 | CP007457 |
| 30 | B. pullorum | DSM 20433 = JCM 1214 | NZ_JDUI01000001 | D86196 |
| 31 | B. reuteri | DSM 23975 | NZ_JDUW01000002 | NZ_JDUW01000049 |
| 32 | B. ruminantium | DSM 6489 = JCM 8222 | NZ_JHWQ01000003 | D86197 |
| 33 | B. saeculare | DSM 6531 | JGZM01000001 | D89328 |
| 34 | B. scardovii | JCM 12489 | AP012331 | AP012331 |
| 35 | B. stellenboschense | DSM 23968 | NZ_JGZP01000019 | JGZP01000012 |
| 36 | B. stercoris | JCM 15918 | JGZQ01000008 | NZ_JDUX01000017 |
| 37 | B. subtile | DSM 20096 | NZ_AUFH01000005 | D89378 |
| 38 | B. thermacidophilum subsp. porcinum | LMG 21689 | JGZS01000003 | NZ_JGZS01000003 |
| 39 | B. thermophilum | RBL67 | CP004346 | DQ340557 |
| 40 | B. tsurumiense | DSM 17777 = OMB115 | NZ_AUCL01000007 | AB241106 |
| Sample ID | Sequence Number a (16S) | OTU Number b (16S) | Sequence Number (groEL) | OTU Number (groEL) |
|---|---|---|---|---|
| H1 | 8774 | 3187 | 8583 | 2044 |
| H2 | 29297 | 1566 | 20519 | 3050 |
| H3 | 26457 | 1511 | 23778 | 2755 |
| H4 | 28524 | 2632 | 18206 | 3176 |
| H5 | 18197 | 970 | 15605 | 2536 |
| H6 | 19625 | 2045 | 15297 | 3943 |
| H7 | 13352 | 1137 | 14950 | 3457 |
| H8 | 37031 | 3429 | 19550 | 4349 |
| R1 | 13986 | 1351 | 9341 | 736 |
| R2 | 9721 | 1279 | 9385 | 815 |
| R3 | 13187 | 1462 | 34297 | 1665 |
| R4 | 16230 | 1404 | 27903 | 1545 |
| R5 | 43429 | 936 | 17599 | 967 |
| R6 | 19969 | 596 | 19369 | 1060 |
| R7 | 18249 | 723 | 19835 | 1111 |
| R8 | 17262 | 930 | 13146 | 870 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Lu, W.; Wang, L.; Pan, M.; Zhang, H.; Zhao, J.; Chen, W. Assessment of Bifidobacterium Species Using groEL Gene on the Basis of Illumina MiSeq High-Throughput Sequencing. Genes 2017, 8, 336. https://doi.org/10.3390/genes8110336
Hu L, Lu W, Wang L, Pan M, Zhang H, Zhao J, Chen W. Assessment of Bifidobacterium Species Using groEL Gene on the Basis of Illumina MiSeq High-Throughput Sequencing. Genes. 2017; 8(11):336. https://doi.org/10.3390/genes8110336
Chicago/Turabian StyleHu, Lujun, Wenwei Lu, Linlin Wang, Mingluo Pan, Hao Zhang, Jianxin Zhao, and Wei Chen. 2017. "Assessment of Bifidobacterium Species Using groEL Gene on the Basis of Illumina MiSeq High-Throughput Sequencing" Genes 8, no. 11: 336. https://doi.org/10.3390/genes8110336




