Dystrophin Dp116: A yet to Be Investigated Product of the Duchenne Muscular Dystrophy Gene
Abstract
:1. Introduction
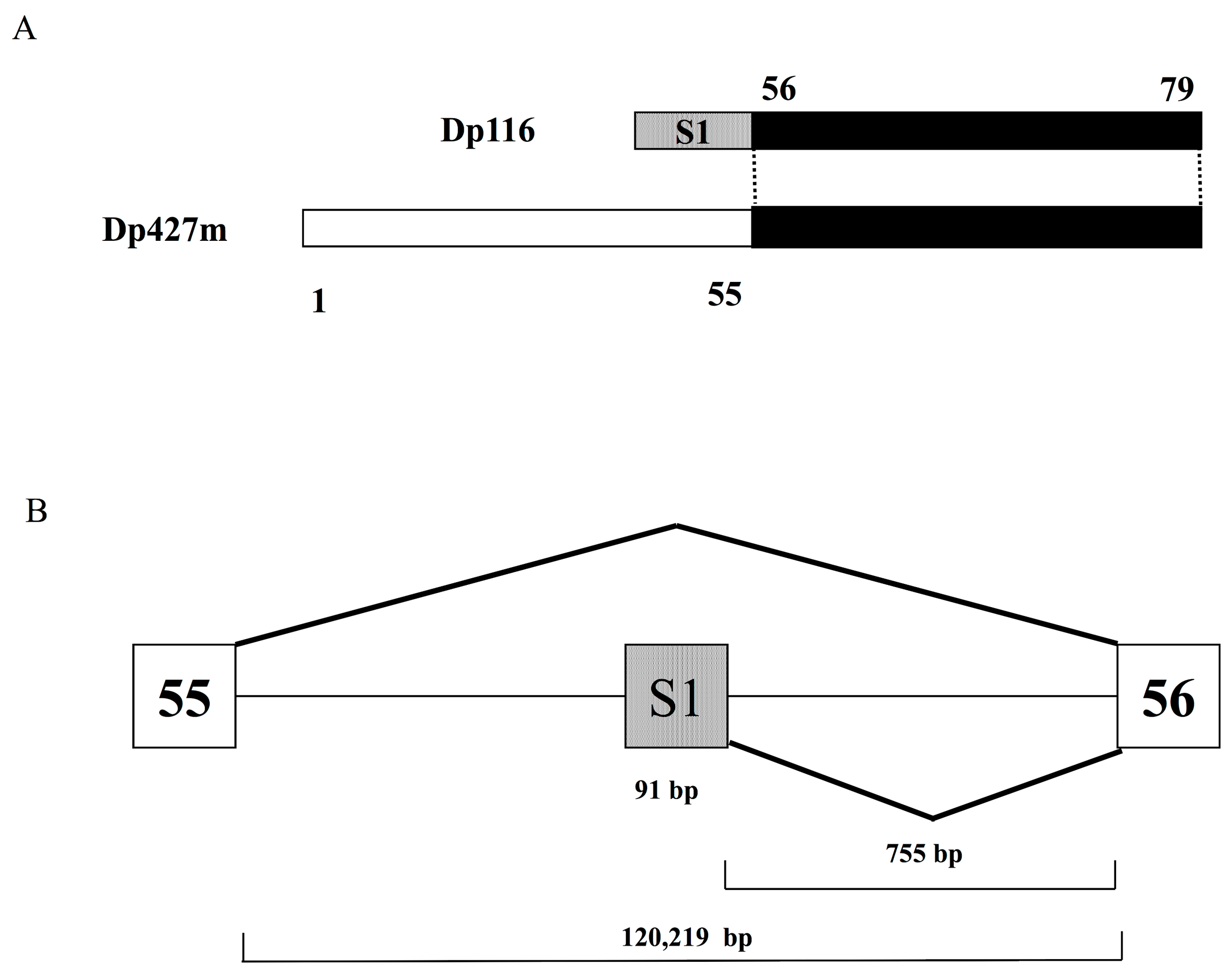
2. The Structure of Dp116 cDNA and Its Relationship to the DMD Gene
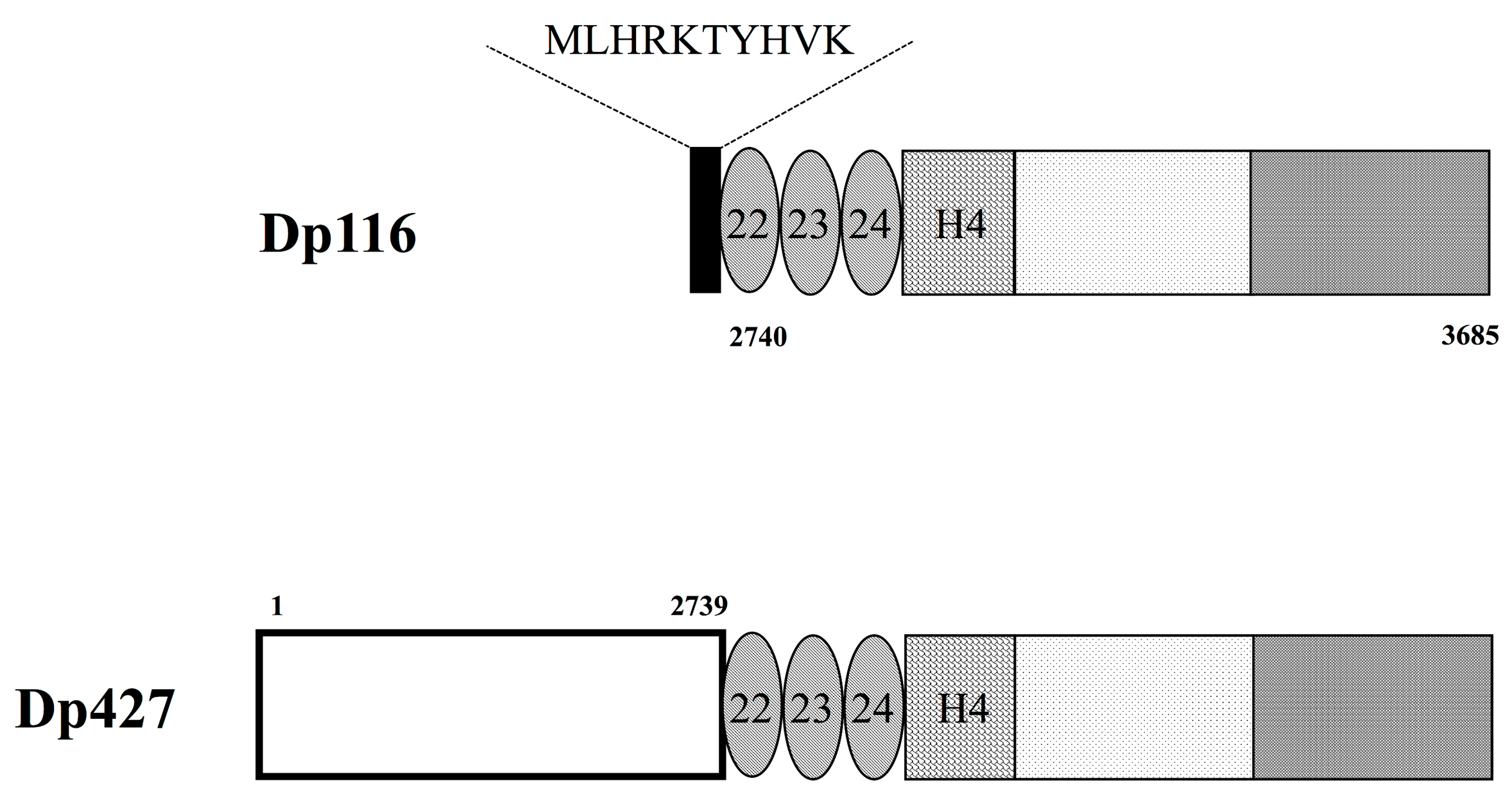
3. Characteristics of Dp116 Protein
4. Characterization of Dp116
4.1. Non-Human Studies of Dp116
4.2. Up113, an Autosomal Homologue of Dp116
4.3. Transgenic Expression of Human Dp116 in Mdx Mice
5. Clinical Findings Associated with Dp116
6. Dp118, a New Candidate Product from DMD Exon S1
6.1. Dp118 Produced by Activation of an Alternative Translation Initiation Site
6.2. Characterization of Dp118
6.3. Future Studies to Confirm the Presence of Dp118
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bushby, K.; Finkel, R.; Birnkrant, D.J.; Case, L.E.; Clemens, P.R.; Cripe, L.; Kaul, A.; Kinnett, K.; McDonald, C.; Pandya, S.; et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: Diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010, 9, 77–93. [Google Scholar] [CrossRef]
- Emery, A.E.H. Confirmation of the diagnosis. In Duchenne Muscular Dystrophy, 2nd ed.; Oxford University Press: Oxford, UK, 1993; Volume 24, pp. 45–79. [Google Scholar]
- Costa, M.F.; Oliveira, A.G.; Feitosa-Santana, C.; Zatz, M.; Ventura, D.F. Red-green color vision impairment in Duchenne muscular dystrophy. Am. J. Hum. Genet. 2007, 80, 1064–1075. [Google Scholar] [CrossRef] [PubMed]
- Pillers, D.A.; Fitzgerald, K.M.; Duncan, N.M.; Rash, S.M.; White, R.A.; Dwinnell, S.J.; Powell, B.R.; Schnur, R.E.; Ray, P.N.; Cibis, G.W.; et al. Duchenne/Becker muscular dystrophy: Correlation of phenotype by electroretinography with sites of dystrophin mutations. Hum. Genet. 1999, 105, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Wood, C.L.; Straub, V.; Guglieri, M.; Bushby, K.; Cheetham, T. Short stature and pubertal delay in Duchenne muscular dystrophy. Arch. Dis. Child. 2016, 101, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Awano, H.; Lee, T.; Takeshima, Y.; Matsuo, M.; Iijima, K. Patients with Duchenne muscular dystrophy are significantly shorter than those with Becker muscular dystrophy, with the higher incidence of short stature in Dp71 mutated subgroup. Neuromuscul. Disord. 2017, in press. [Google Scholar] [CrossRef] [PubMed]
- Muntoni, F.; Torelli, S.; Ferlini, A. Dystrophin and mutations: One gene, several proteins, multiple phenotypes. Lancet Neurol. 2003, 2, 731–740. [Google Scholar] [CrossRef]
- Pillers, D.A.; Bulman, D.E.; Weleber, R.G.; Sigesmund, D.A.; Musarella, M.A.; Powell, B.R.; Murphey, W.H.; Westall, C.; Panton, C.; Becker, L.E.; et al. Dystrophin expression in the human retina is required for normal function as defined by electroretinography. Nat. Genet. 1993, 4, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Ricotti, V.; Jagle, H.; Theodorou, M.; Moore, A.T.; Muntoni, F.; Thompson, D.A. Ocular and neurodevelopmental features of Duchenne muscular dystrophy: A signature of dystrophin function in the central nervous system. Eur. J. Hum. Genet. 2016, 24, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Doorenweerd, N.; Straathof, C.S.; Dumas, E.M.; Spitali, P.; Ginjaar, I.B.; Wokke, B.H.; Schrans, D.G.; van den Bergen, J.C.; van Zwet, E.W.; Webb, A.; et al. Reduced cerebral gray matter and altered white matter in boys with Duchenne muscular dystrophy. Ann. Neurol. 2014, 76, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Doorenweerd, N.; Dumas, E.M.; Ghariq, E.; Schmid, S.; Straathof, C.S.M.; Roest, A.A.W.; Wokke, B.H.; van Zwet, E.W.; Webb, A.G.; Hendriksen, J.G.M.; et al. Decreased cerebral perfusion in Duchenne muscular dystrophy patients. Neuromuscul. Disord. 2017, 27, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Tadayoni, R.; Rendon, A.; Soria-Jasso, L.E.; Cisneros, B. Dystrophin Dp71: The smallest but multifunctional product of the Duchenne muscular dystrophy gene. Mol. Neurobiol. 2012, 45, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Byers, T.J.; Lidov, H.G.; Kunkel, L.M. An alternative dystrophin transcript specific to peripheral nerve. Nat. Genet. 1993, 4, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Schofield, J.N.; Blake, D.J.; Simmons, C.; Morris, G.E.; Tinsley, J.M.; Davies, K.E.; Edwards, Y.H. Apo-dystrophin-1 and apo-dystrophin-2, products of the Duchenne muscular dsytrophy locus: Expression during mouse embryogenesis and in cultured cell lines. Hum. Mol. Genet. 1994, 3, 1309–1316. [Google Scholar] [PubMed]
- Mizuno, Y.; Yoshida, M.; Yamamoto, H.; Hirai, S.; Ozawa, E. Distribution of dystrophin isoforms and dystrophin-associated proteins 43DAG (A3a) and 50DAG (A2) in various monkey tissues. J. Biochem. 1993, 114, 936–941. [Google Scholar] [CrossRef] [PubMed]
- 1Blitzblau, R.; Storer, E.K.; Jacob, M.H. Dystrophin and utrophin isoforms are expressed in glia, but not neurons, of the avian parasympathetic ciliary ganglion. Brain Res. 2008, 1218, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Vita, G.; Mazzeo, A.; Muglia, U.; Girlanda, P.; Toscano, A.; Rodolico, C.; Migliorato, A. Dp116, talin, vinculin and vimentin immunoreactivities following nerve transection. Neuroreport 1998, 9, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, K.; Yamada, H.; Shimizu, T.; Campbell, K.P. Differential expression of dystrophin, utrophin and dystrophin-associated proteins in peripheral nerve. FEBS Lett. 1993, 334, 281–285. [Google Scholar] [CrossRef]
- Fabbrizio, E.; Latouche, J.; Rivier, F.; Hugon, G.; Mornet, D. Re-evaluation of the distributions of dystrophin and utrophin in sciatic nerve. Biochem. J. 1995, 312, 309–314. [Google Scholar] [CrossRef]
- Cai, H.; Erdman, R.A.; Zweier, L.; Chen, J.; Shaw, J.H., IV; Baylor, K.A.; Stecker, M.M.; Carey, D.J.; Chan, Y.M. The sarcoglycan complex in Schwann cells and its role in myelin stability. Exp. Neurol. 2007, 205, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Imamura, M.; Araishi, K.; Noguchi, S.; Ozawa, E. A sarcoglycan-dystroglycan complex anchors Dp116 and utrophin in the peripheral nervous system. Hum. Mol. Genet. 2000, 9, 3091–3100. [Google Scholar] [CrossRef] [PubMed]
- Hnia, K.; Hugon, G.; Masmoudi, A.; Mercier, J.; Rivier, F.; Mornet, D. Effect of beta-dystroglycan processing on utrophin/Dp116 anchorage in normal and mdx mouse Schwann cell membrane. Neuroscience 2006, 141, 607–620. [Google Scholar] [CrossRef] [PubMed]
- 2Scherer, S.S.; Arroyo, E.J. Recent progress on the molecular organization of myelinated axons. J. Peripher. Nerv. Syst. 2002, 7, 1–12. [Google Scholar] [CrossRef]
- Sherman, D.L.; Wu, L.M.; Grove, M.; Gillespie, C.S.; Brophy, P.J. Drp2 and periaxin form Cajal bands with dystroglycan but have distinct roles in Schwann cell growth. J. Neurosci. 2012, 32, 9419–9428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, D.E.; Sherman, D.L.; Brophy, P.J.; Froehner, S.C. The ABCA1 cholesterol transporter associates with one of two distinct dystrophin-based scaffolds in Schwann cells. Glia 2008, 56, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Romo-Yanez, J.; Montanez, C.; Salazar-Olivo, L.A. Dystrophins and DAPs are expressed in adipose tissue and are regulated by adipogenesis and extracellular matrix. Biochem. Biophys. Res. Commun. 2011, 404, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Taghli-Lamallem, O.; Akasaka, T.; Hogg, G.; Nudel, U.; Yaffe, D.; Chamberlain, J.S.; Ocorr, K.; Bodmer, R. Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell 2008, 7, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Guiraud, S.; Davies, K.E. Pharmacological advances for treatment in Duchenne muscular dystrophy. Curr. Opin. Pharmacol. 2017, 34, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Blake, D.J.; Schofield, J.N.; Zuellig, R.A.; Gorecki, D.C.; Phelps, S.R.; Barnard, E.A.; Edwards, Y.H.; Davies, K.E. G-utrophin, the autosomal homologue of dystrophin Dp116, is expressed in sensory ganglia and brain. Proc. Natl. Acad. Sci. USA 1995, 92, 3697–3701. [Google Scholar] [CrossRef] [PubMed]
- Knuesel, I.; Bornhauser, B.C.; Zuellig, R.A.; Heller, F.; Schaub, M.C.; Fritschy, J.M. Differential expression of utrophin and dystrophin in CNS neurons: An in situ hybridization and immunohistochemical study. J. Comp. Neurol. 2000, 422, 594–611. [Google Scholar] [CrossRef]
- Judge, L.M.; Arnett, A.L.; Banks, G.B.; Chamberlain, J.S. Expression of the dystrophin isoform Dp116 preserves functional muscle mass and extends lifespan without preventing dystrophy in severely dystrophic mice. Hum. Mol. Genet. 2011, 20, 4978–4990. [Google Scholar] [CrossRef] [PubMed]
- Labarque, V.; Freson, K.; Thys, C.; Wittevrongel, C.; Hoylaerts, M.F.; De Vos, R.; Goemans, N.; Van Geet, C. Increased Gs signalling in platelets and impaired collagen activation, due to a defect in the dystrophin gene, result in increased blood loss during spinal surgery. Hum. Mol. Genet. 2008, 17, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, S.; Tsuruta, K.; Kurihara, T.; Ono, S.; Morotomi, Y.; Inoue, K.; Matsukura, S. Posterior tibial somatosensory evoked potentials in Duchenne-type progressive muscular dystrophy. Electroencephalogr. Clin. Neurophysiol. 1986, 64, 525–527. [Google Scholar] [CrossRef]
- Kraus, D.; Wong, B.L.; Horn, P.S.; Kaul, A. Constipation in Duchenne muscular dystrophy: Prevalence, diagnosis, and treatment. J. Pediatr. 2016, 171, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Latimer, R.; Street, N.; Conway, K.C.; James, K.; Cunniff, C.; Oleszek, J.; Fox, D.; Ciafaloni, E.; Westfield, C.; Paramsothy, P.; et al. Secondary conditions among males with Duchenne or Becker muscular dystrophy. J. Child Neurol. 2017, 32, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Lo Cascio, C.M.; Goetze, O.; Latshang, T.D.; Bluemel, S.; Frauenfelder, T.; Bloch, K.E. Gastrointestinal dysfunction in patients with Duchenne muscular dystrophy. PLoS ONE 2016, 11, e0163779. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.P.; Garcia, C.A.; Chamberlain, J.S.; Angelini, C.; Lupski, J.R.; Fenwick, R. Is the carboxy-terminus of dystrophin required for membrane association? A novel, severe case of Duchenne muscular dystrophy. Ann. Neurol. 1991, 30, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, T.R.; Ellis, J.M.; Mountford, R.C.; Appleton, R.E.; Morris, G.E. A truncated dystrophin lacking the C-terminal domains is localized at the muscle membrane. Am. J. Hum. Genet. 1992, 50, 508–514. [Google Scholar] [PubMed]
- Recan, D.; Chafey, P.; Leturcq, F.; Hugnot, J.P.; Vincent, N.; Tome, F.; Collin, H.; Simon, D.; Czernichow, P.; Nichoson, L.V.B.; et al. Are cysteine-rich and COOH-terminal domains of dystrophin critical for sarcolemmal localization? J. Clin. Investig. 1992, 89, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Wibawa, T.; Takeshima, Y.; Mitsuyoshi, I.; Wada, H.; Surono, A.; Nakamura, H.; Matsuo, M. Complete skipping of exon 66 due to novel mutations of the dystrophin gene was identified in two Japanese families of Duchenne muscular dystrophy with severe mental retardation. Brain Dev. 2000, 22, 107–112. [Google Scholar] [CrossRef]
- Pane, M.; Messina, S.; Bruno, C.; D’Amico, A.; Villanova, M.; Brancalion, B.; Sivo, S.; Bianco, F.; Striano, P.; Battaglia, D.; et al. Duchenne muscular dystrophy and epilepsy. Neuromuscul. Disord. 2013, 23, 313–315. [Google Scholar] [CrossRef] [PubMed]
- Leiden Muscular Dystrophy pages. Available online: http://www.dmd.nl/DMD_home.html (accessed on 11 June 2017).
- Chiurazzi, P.; Schwartz, C.E.; Gecz, J.; Neri, G. XLMR genes: Update 2007. Eur. J. Hum. Genet. 2008, 16, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Felisari, G.; Martinelli Boneschi, F.; Bardoni, A.; Sironi, M.; Comi, G.P.; Robotti, M.; Turconi, A.C.; Lai, M.; Corrao, G.; Bresolin, N. Loss of Dp140 dystrophin isoform and intellectual impairment in Duchenne dystrophy. Neurology 2000, 55, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.J.; Betts, G.A.; Maroulis, S.; Gilissen, C.; Pedersen, R.L.; Mowat, D.R.; Johnston, H.M.; Buckley, M.F. Dystrophin gene mutation location and the risk of cognitive impairment in Duchenne muscular dystrophy. PLoS ONE 2010, 5, e8803. [Google Scholar] [CrossRef] [PubMed]
- Wingeier, K.; Giger, E.; Strozzi, S.; Kreis, R.; Joncourt, F.; Conrad, B.; Gallati, S.; Steinlin, M. Neuropsychological impairments and the impact of dystrophin mutations on general cognitive functioning of patients with Duchenne muscular dystrophy. J. Clin. Neurosci. 2011, 18, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Ricotti, V.; Mandy, W.P.; Scoto, M.; Pane, M.; Deconinck, N.; Messina, S.; Mercuri, E.; Skuse, D.H.; Muntoni, F. Neurodevelopmental, emotional, and behavioural problems in Duchenne muscular dystrophy in relation to underlying dystrophin gene mutations. Dev. Med. Child Neurol. 2016, 58, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Mazzeo, A.; Rodolico, C.; Monici, M.C.; Migliorato, A.; Aguennouz, M.; Vita, G. Perineurium talin immunoreactivity decreases in diabetic neuropathy. J. Neurol. Sci. 1997, 146, 7–11. [Google Scholar] [CrossRef]
- Sanders, A.E.; Jain, D.; Sofer, T.; Kerr, K.F.; Laurie, C.C.; Shaffer, J.R.; Marazita, M.L.; Kaste, L.M.; Slade, G.D.; Fillingim, R.B.; et al. GWAS identifies new loci for painful temporomandibular disorder: Hispanic Community Health Study/Study of Latinos. J. Dent. Res. 2017, 96, 277–284. [Google Scholar] [CrossRef] [PubMed]
- BLAST. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 11 June 2017).
- Kozak, M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984, 12, 857–872. [Google Scholar] [CrossRef] [PubMed]
- TIS miner. Available online: http://dnafsminer.bic.nus.edu.sg/Tis.html (accessed on 11 June 2017).
- Liu, H.; Wong, L. Data mining tools for biological sequences. J. Bioinform. Comput. Biol. 2003, 1, 139–167. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Noriega, A.; Michalak, C.; Cervantes-Roldan, R.; Gomez-Romero, V.; Leon-Del-Rio, A. Two translation initiation codons direct the expression of annexin VI 64kDa and 68kDa isoforms. Mol. Genet. Metab. 2016, 119, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Tan, S.; Hermanowski, J.; Bohm, S.; Pacheco, S.; McCauley, J.M.; Greener, M.J.; Hinits, Y.; Hughes, S.M.; Sharpe, P.T.; et al. The dystrotelin, dystrophin and dystrobrevin superfamily: New paralogues and old isoforms. BMC Genomics 2007, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Tarnow, P.; Schoneberg, T.; Krude, H.; Gruters, A.; Biebermann, H. Mutationally induced disulfide bond formation within the third extracellular loop causes melanocortin 4 receptor inactivation in patients with obesity. J. Biol. Chem. 2003, 278, 48666–48673. [Google Scholar] [CrossRef] [PubMed]
- Hanggi, E.; Grundschober, A.F.; Leuthold, S.; Meier, P.J.; St-Pierre, M.V. Functional analysis of the extracellular cysteine residues in the human organic anion transporting polypeptide, OATP2B1. Mol. Pharmacol. 2006, 70, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Judge, L.M.; Haraguchiln, M.; Chamberlain, J.S. Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. J. Cell Sci. 2006, 119, 1537–1546. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuo, M.; Awano, H.; Matsumoto, M.; Nagai, M.; Kawaguchi, T.; Zhang, Z.; Nishio, H. Dystrophin Dp116: A yet to Be Investigated Product of the Duchenne Muscular Dystrophy Gene. Genes 2017, 8, 251. https://doi.org/10.3390/genes8100251
Matsuo M, Awano H, Matsumoto M, Nagai M, Kawaguchi T, Zhang Z, Nishio H. Dystrophin Dp116: A yet to Be Investigated Product of the Duchenne Muscular Dystrophy Gene. Genes. 2017; 8(10):251. https://doi.org/10.3390/genes8100251
Chicago/Turabian StyleMatsuo, Masafumi, Hiroyuki Awano, Masaaki Matsumoto, Masashi Nagai, Tatsuya Kawaguchi, Zhujun Zhang, and Hisahide Nishio. 2017. "Dystrophin Dp116: A yet to Be Investigated Product of the Duchenne Muscular Dystrophy Gene" Genes 8, no. 10: 251. https://doi.org/10.3390/genes8100251




