The Complete Chloroplast Genome of the Hare’s Ear Root, Bupleurum falcatum: Its Molecular Features
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sequencing, Assembly, and Validation of the B. falcatum Chloroplast Genome
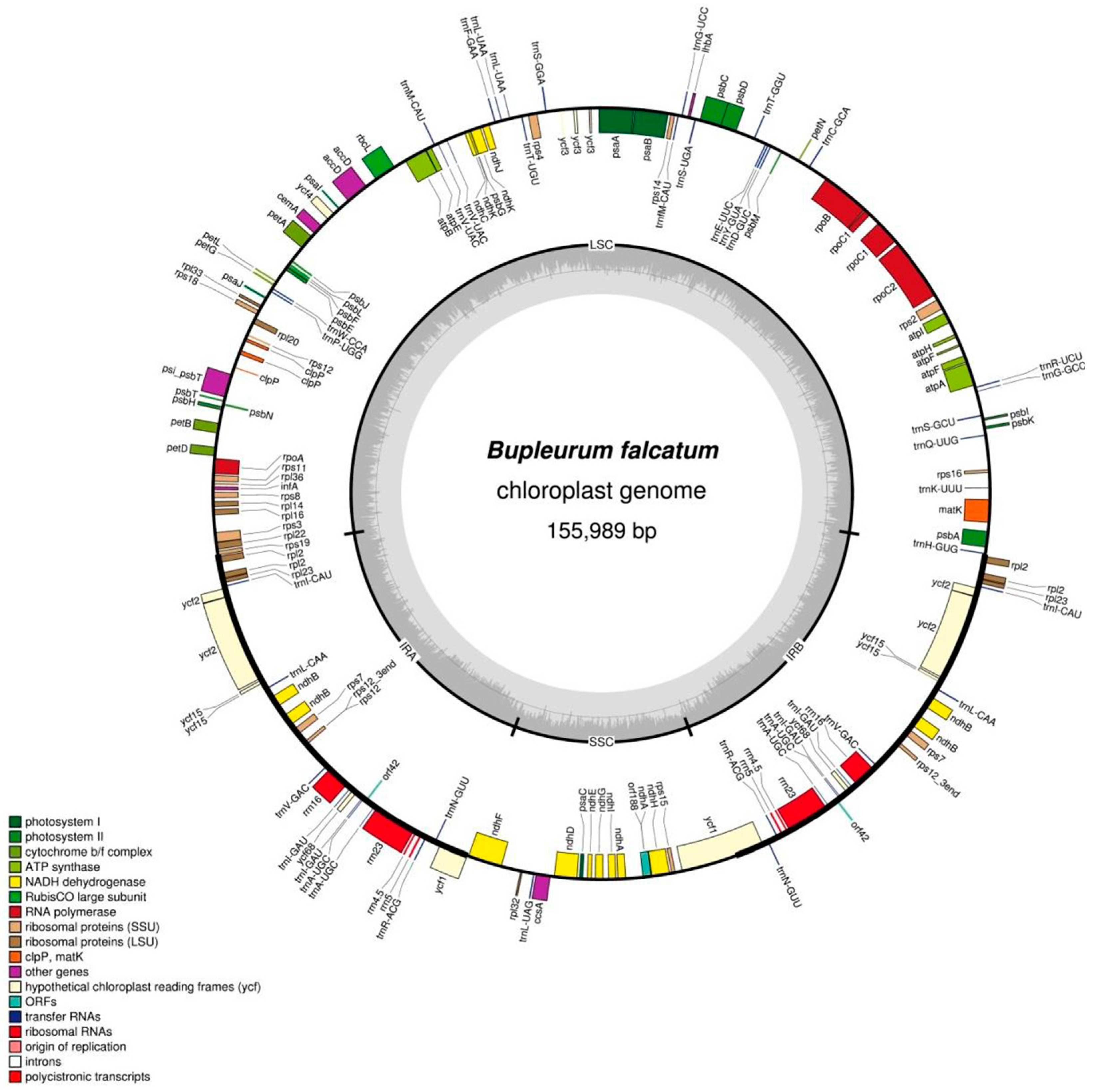
2.2. Physical Features of the B. falcatum Chloroplast Genome
2.3. Tandem Repeat Sequences
3. Experimental Section
3.1. Plant and DNA Sample Preparation
3.2. Chloroplast Genome Sequencing, Assembly, and Validation
3.3. Chloroplast Genome Annotation, Codon Usage and Repeat Sequence Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| LSC | Large Sequence Copy |
| IR | Inverted Repeat |
| SSC | Small Sequence Copy |
| TR | Tandem Repeats |
| CP | Chloroplast |
References
- Chloroplast. Available online: http://en.wikipedia.org/wiki/Chloroplast_DNA (accessed on 7 December 2015).
- Neuhaus, H.E.; Emes, M.J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwonse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef] [PubMed]
- Ris, H.; Plaut, W. Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J. Cell Biol. 1962, 13, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Birky, C.W., Jr. Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc. Natl. Acad. Sci. USA 1995, 95, 11331–11338. [Google Scholar] [CrossRef]
- Ilona, M. Non-nuclear genes and their inheritance. Nat. Educ. 2008, 1, 135. [Google Scholar]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequence to choose noncoding regions for phylogenetic studies in Angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M. The chloroplast genome. Plant Mol. Biol. 1992, 19, 149–168. [Google Scholar] [CrossRef] [PubMed]
- Gary, M.W. The evolutionary origins of organelles. Trends Genet. 1989, 5, 294–299. [Google Scholar] [CrossRef]
- Leseberg, C.H.; Duvall, M.R. The complete chloroplast genome of coix lacryma-jobi and a comparative molecular evolutionary analysis of plastomes in cereals. J. Mol. Evol. 2009, 69, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Gary, M.W.; Sankoff, D.; Cedergren, R.J. On the evolutionary descent of organisms and organelles: A global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984, 12, 5837–5852. [Google Scholar] [CrossRef]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The complete chloroplast genome sequence of Pelargonium x hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Thompson, W.F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 1982, 29, 537–550. [Google Scholar] [CrossRef]
- Palmer, J.D. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Morden, C.W.; Ems, S.C.; Palmer, J.D. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: Loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J. Mol. Evol. 1992, 35, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.L.; Jansen, R.K.; Chumley, T.W.; Kim, K.J. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 2007, 24, 1161–1180. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; dePamphilis, C.W.; Leebens-Meck, J.; Muller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Morgante, M.; McDevitt, R.; Vendramin, G.G.; Rafalski, J.A. Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. USA 1995, 92, 7759–7763. [Google Scholar] [CrossRef] [PubMed]
- Bock, R.; Khan, M.S. Taming plastids for a green future. Trends Biotechnol. 2004, 22, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Lee, H.L. Complete chloroplast genome sequences from Korean Ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ma, P.F.; Wen, J.; Yi, T.S. Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications. PLoS ONE 2013, 8. [Google Scholar]
- Lee, B.B.; Shim, I.; Lee, H.J.; Hahm, D.H. Effect of Bupleurum falcatum on the stress-induced impairment of spatial working memory in rats. Biol. Pharm. Bull. 2009, 32, 1392–1398. [Google Scholar] [PubMed]
- Lee, B.B.; Yun, H.Y.; Shim, I.S.; Lee, H.J.; Hahm, D.H. Bupleurum falcatum prevents depression and anxiety-like behaviors in rats exposed to repeated restraint stress. J. Microbiol. Biotechnol. 2012, 22, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, H.; Chino, A.; Hirasaki, Y.; Ueda, K.; Raimura, M.; Namiki, T. A valid approach in refractory glossodynia: single-institution 5-year experience treating with Japanese traditional herbal (kampo) medicine. Evid. Based Complement. Altern. Med. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Bang, O.S.; Kim, J.H. Screening of Stat3 inhibitory effects of Korean herbal medicines in the A549 human lung cancer cell line. Integr. Med. Res. 2014, 3, 67–73. [Google Scholar] [CrossRef]
- Zhao, Y.; Yin, J.; Guo, H.; Zhang, Y.; Xiao, W.; Sun, C.; Wu, J.; Qu, X.; Yu, J.; Wang, X.; et al. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front. Plant Sci. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Angiosperm Phylogeny Group III. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG. Bot. J. Linn. Soc. 2009, 161, 105–121. [Google Scholar]
- Sugita, M.; Kato, A.; Shimada, H.; Sugiura, M. Sequence analysis of the junctions between a large inverted repeat and single-copy regions in tobacco chloroplast DNA. Mol. Gen. Genet. 1984, 194, 200–205. [Google Scholar] [CrossRef]
- Konishi, T.; Shinohara, K.; Yamada, K.; Sasaki, Y. Acetyl-CoA carboxylase in higher plants: Most plants other than Gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 1996, 37, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, J.; Shimda, H.; Whittier, R.; Ishibashi, T.; Sakamoto, M.; Mori, M.; Kondo, C.; Honji, Y.; Sun, C.R.; Meng, B.Y.; et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: Itermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 1989, 217, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.M.; Neckermann, K.; Igloi, G.L.; Kossel, H. Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995, 251, 614–628. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, Y.; Isono, K.; Kojima, T.; Endo, A.; Hanaoka, M.; Shiina, T.; Terachi, T.; Utsugi, S.; Murata, M.; Mori, N.; et al. Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol. Genet. Genom. 2002, 266, 740–746. [Google Scholar]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethyl-ammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lee, S.C.; Lee, J.K.; Lee, H.O.; Joh, H.J.; Kim, N.H.; Park, H.S.; Yang, T.J. Comprehensive survey of genetic diversity in chloroplast genome and 45S nrDNAs within Panax ginseng Species. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jensen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acid Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
| Compositional Category | Features in Chloroplast |
|---|---|
| CP genome length (nt) | 155,989 |
| LSC length (nt) | 85,912 |
| SSC length (nt) | 17,517 |
| IR length (Ira/b) (nt) | 52,560 |
| AT/GC contents (%) | 62.34/37.66 |
| No. of total/unique genes | 129/111 |
| Genic occupancy (nt) | 88,065 |
| Intergenic occupancy (nt) | 67,924 |
| Protein-coding genes | 78 |
| Transfer RNAs (tRNAs) | 29 |
| Ribosomal RNAs (rRNAs) | 4 |
| No. of genes duplicated in IR regions | 16 |
| Total number of genes with intron(s) | 17 |
| Gene(s) with single intron | 14 |
| Gene(s) with multiple introns | 3 |
| tRNAs with intron(s) | 5 |
| Category of Gene Group | Group of Genes | Name of Genes |
|---|---|---|
| Self replication | Ribosomal RNAs | 16S (rrn16)(x2), 23S (rrn23)(x2) |
| 4.5S (rrn4.5)(x2), 5S (rrn5)(x2) | ||
| trnH-GUG, trnK-UUU, trnQ-UUG | ||
| trnS-GCU, trnG-UCC †, trnR-UCU | ||
| trnC-GCA, trnD-GUC, trnY-GUA | ||
| trnE-UUC, trnT-GGU, trnS-UGA | ||
| trnfM-CAU, trnS-GGA, trnT-UGU | ||
| trnL-UAA †, trnF-GAA, trnV-UAC † | ||
| Transfer RNAs | trnM-CAU, trnW-CCA, trnP-UGG, trnI-CAU(x2), trnL-CAA(x2), trnV-GAC(x2), trnI-GAU(x2) †, trnA-UGC(x2) †, trnR-ACG(x2), trnN-GUU(x2), trnL-UAG | |
| Small subunit of ribosome | rps2, rps3, rps4, rps7(x2), rps8, rps11, rps12(x2, part) †, rps14, rps15, rps16 †, rps18, rps19 | |
| Large subunit of ribosome | rpl2(x2) †, rpl14, rpl16 †, rpl20, rpl22, rpl23(x2), rpl32, rpl33, rpl36 | |
| RNA polymerase | rpoA, rpoB, rpoC1 †, rpoC2 | |
| Photosynthesis | NADH- | ndhA †, ndhB(x2) †, ndhC, ndhD, ndhE |
| dehydrogenase | ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3 #, | |
| Photosystem II | lhbA, psbA, psbC, psbD, psbE, psbF | |
| psbH, psbI, psbJ, psbK, psbL, psbM | ||
| psbN, psi-psbT, psbT | ||
| Cytochrome b/f | petA, petB †, petD †, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF †, atpH, atpI | |
| Rubisco | rbcL | |
| Other genes | infA, matK, clpP #, cemA, ccsA | |
| Unknown function | ORFs ¥ | ycf1(x2, part), ycf2(x2), ycf4 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, D.-H.; Lee, J.-H.; Kang, S.-H.; Ahn, B.-O.; Kim, C.-K. The Complete Chloroplast Genome of the Hare’s Ear Root, Bupleurum falcatum: Its Molecular Features. Genes 2016, 7, 20. https://doi.org/10.3390/genes7050020
Shin D-H, Lee J-H, Kang S-H, Ahn B-O, Kim C-K. The Complete Chloroplast Genome of the Hare’s Ear Root, Bupleurum falcatum: Its Molecular Features. Genes. 2016; 7(5):20. https://doi.org/10.3390/genes7050020
Chicago/Turabian StyleShin, Dong-Ho, Jeong-Hoon Lee, Sang-Ho Kang, Byung-Ohg Ahn, and Chang-Kug Kim. 2016. "The Complete Chloroplast Genome of the Hare’s Ear Root, Bupleurum falcatum: Its Molecular Features" Genes 7, no. 5: 20. https://doi.org/10.3390/genes7050020
APA StyleShin, D.-H., Lee, J.-H., Kang, S.-H., Ahn, B.-O., & Kim, C.-K. (2016). The Complete Chloroplast Genome of the Hare’s Ear Root, Bupleurum falcatum: Its Molecular Features. Genes, 7(5), 20. https://doi.org/10.3390/genes7050020






