Genome-Wide Identification and Characterization of Long Non-Coding RNAs from Mulberry (Morus notabilis) RNA-seq Data
Abstract
:1. Introduction
2. Experimental Section
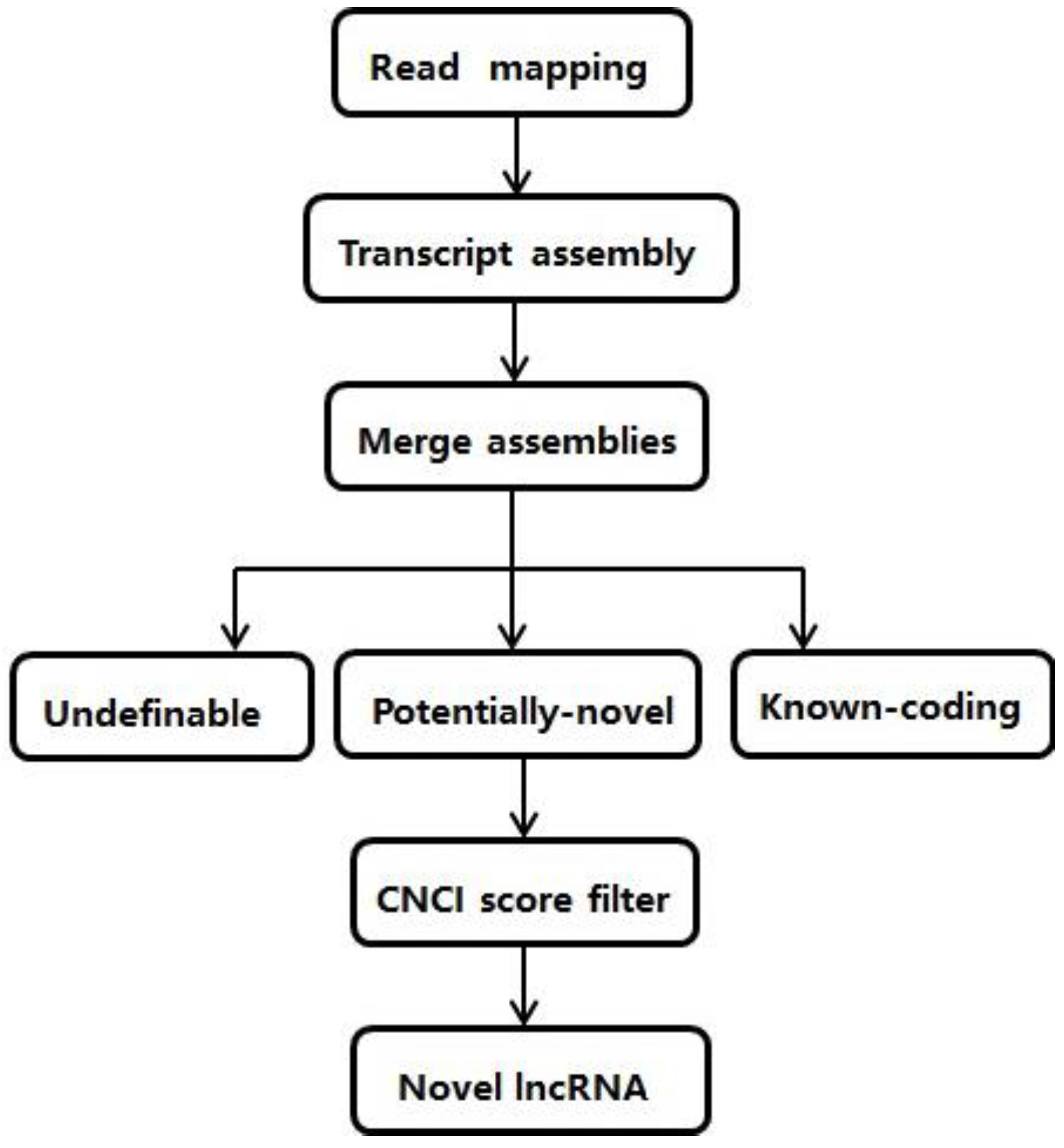
2.1. The Pipeline to Identify lncRNAs from RNA-seq Data
2.2. Calculation of lncRNA Conservation
2.3. Expression Profiles of Tissue Specific lncRNAs and Functional Predictions
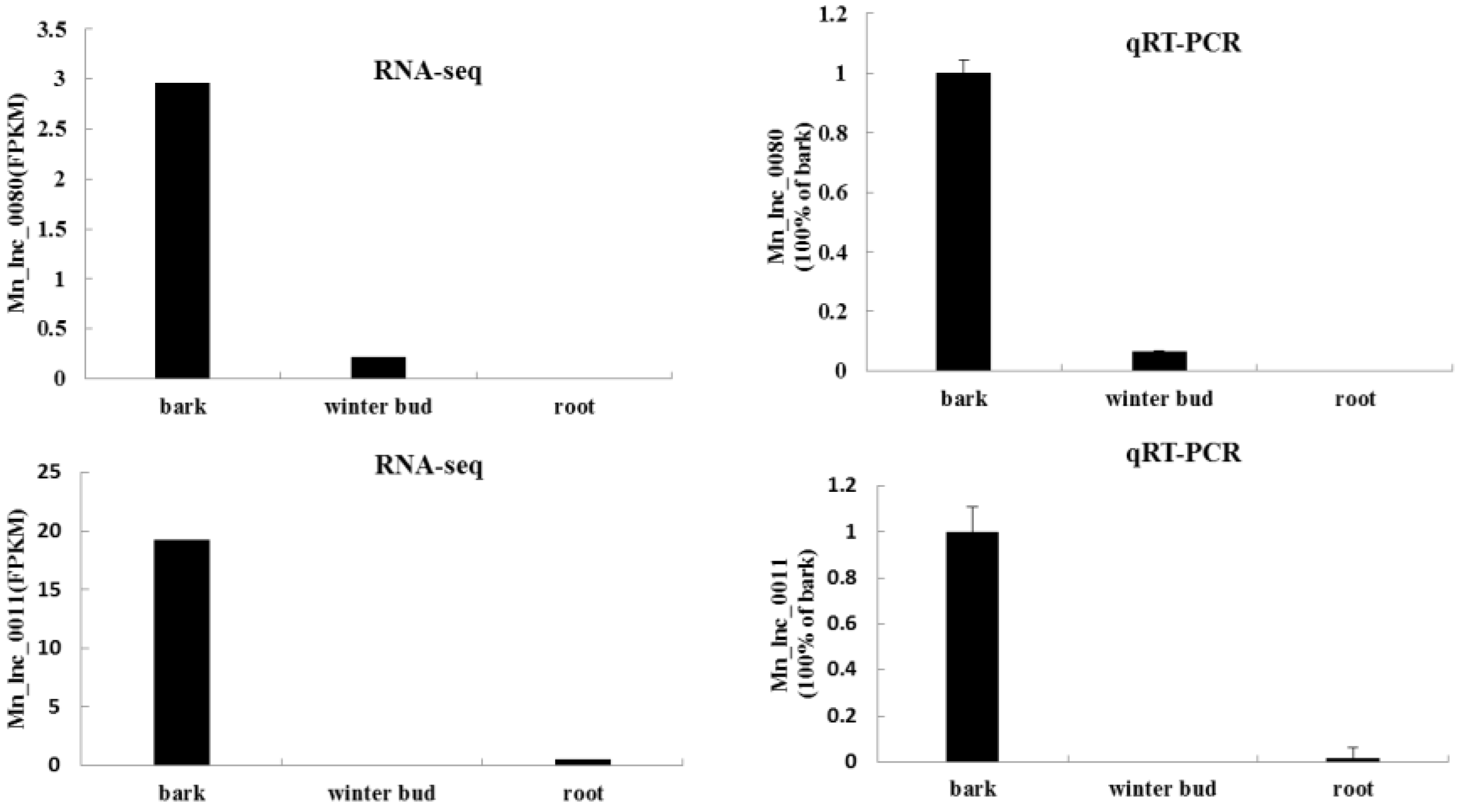
2.4. qRT-PCR Analysis of lncRNAs
3. Results
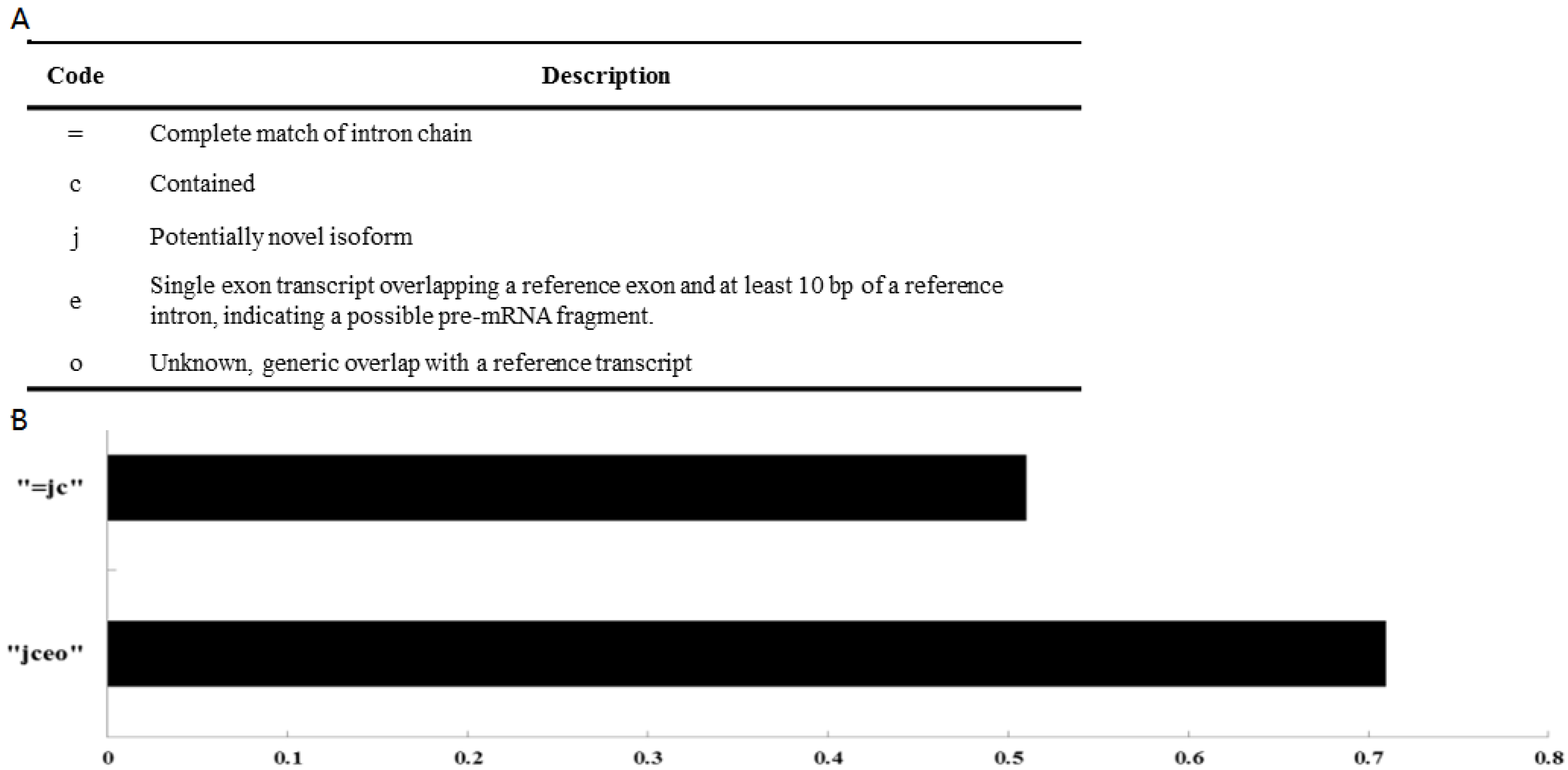
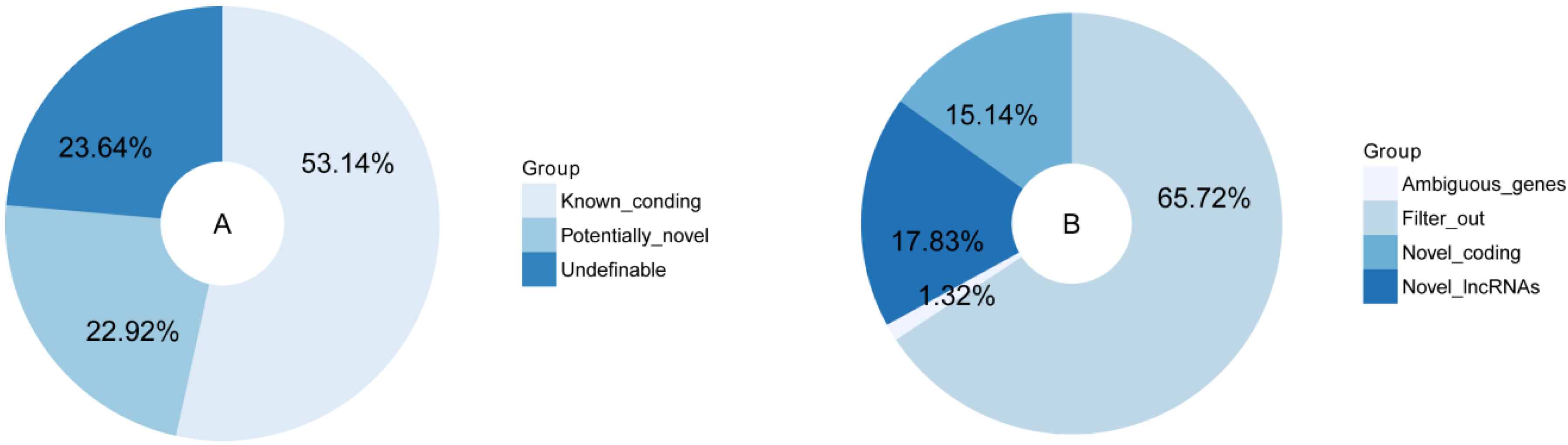
3.1. Transcripts Reconstruction and Identification of Novel lncRNAs
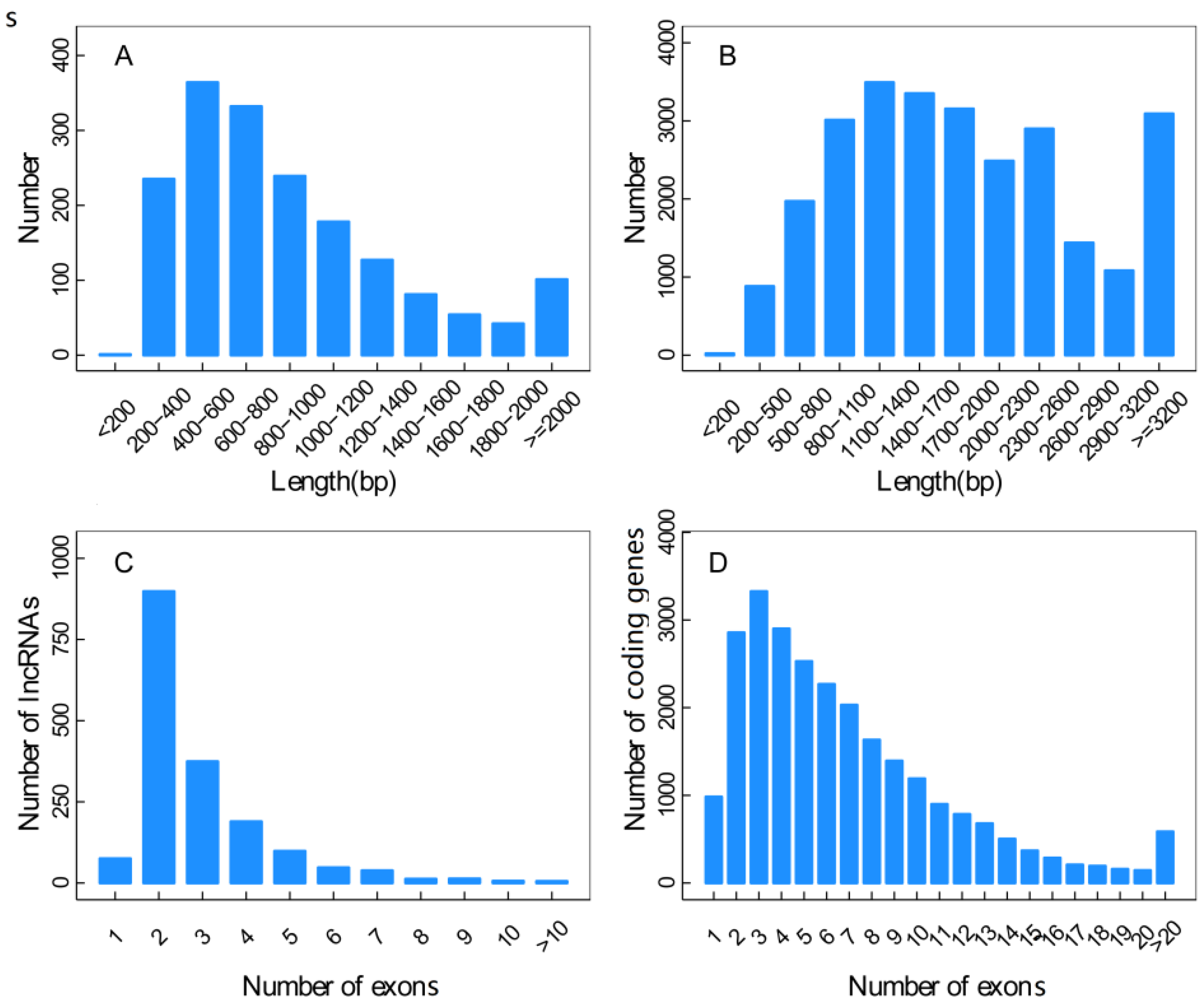
3.2. Characterization of the Novel lncRNAs
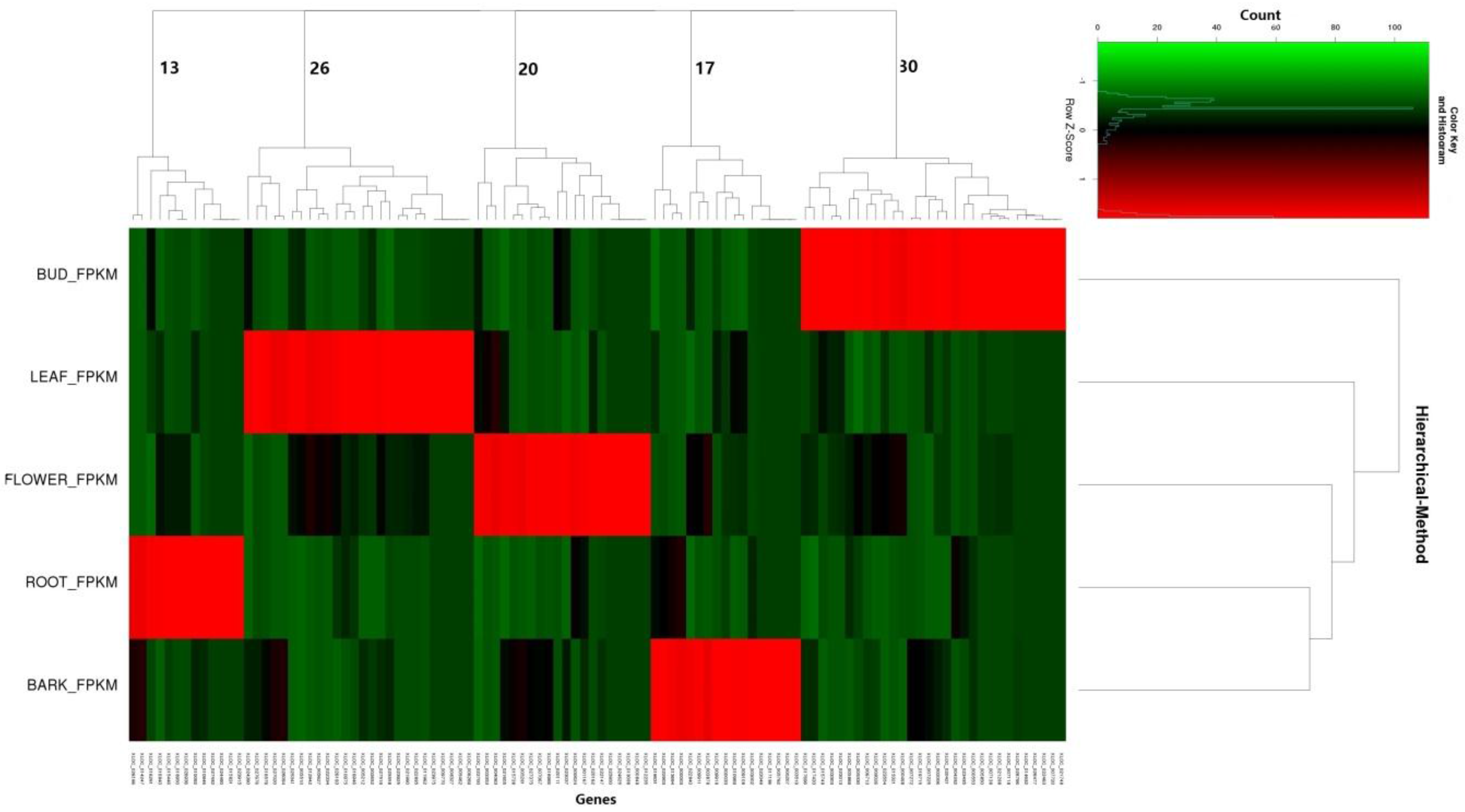
3.3. Expression Profiles of Tissue Specific lncRNAs and Functional Predictions
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Berg, C.C. Moraceae diversity in a global perspective. Biol. Skr. 2005, 55, 423–440. [Google Scholar]
- Nepal, M.P.; Ferguson, C.J. Phylogenetics of Morus (Moraceae) inferred from ITS and trnL-trnF sequence data. Syst. Bot. 2012, 37, 442–450. [Google Scholar] [CrossRef]
- Wang, M.; Gao, L.X.; Wang, J.; Li, J.; Yu, M.; Li, J.; Hou, A. Diels-Alder adducts with PTP1B inhibition from Morus Notabilis. Phytochem. 2015, 109, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Zhang, D.; Qi, X.; Ma, B.; Xiang, Z.; He, N. Identification of the conserved and novel miRNAs in mulberry by high-throughput sequencing. PloS ONE 2014, 9, e104409. [Google Scholar]
- Ning, D.; Lu, B.; Zhang, Y. The processing technology of mulberry series product. China Fruit Veg. Process 2005, 5, 38–40. [Google Scholar]
- Nomura, T.; Fukai, T.; Hano, Y. Chemistry and biological activities of isoprenylated flavonoids from medicinal plants (moraceous plants and Glycyrrhiza species). Stud. Nat. Prod. Chem. 2003, 28, 199–256. [Google Scholar]
- Darias-Martín, J.; Lobo-Rodrigo, G.; Hernández-Cordero, J.; Díaz-Díaz, E.; Díaz-Romero, C. Alcoholic beverages obtained from black mulberry. Food Technol. Biotechnol. 2003, 41, 173–176. [Google Scholar]
- Venkatesh, K.R.; Chauhan, S. Mulberry: Life enhancer. J. Med. Plants Res. 2008, 2, 271–278. [Google Scholar]
- Yang, C.; Fang, X.; Wu, X.; Mao, Y.; Wang, L.; Chen, X. Transcriptional regulation of plant secondary metabolism. J. Integr. Plant Biol. 2012, 54, 703–712. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Zhang, C.; Qi, X.; Zhao, S.; Tao, Z.; Yang, G.; Lee, T.H.; Wang, X.; Cai, Q.; Li, D.; et al. Draft genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Luo, Y.; Jia, L.; Qi, X.; Zeng, Q.; Xiang, Z.; He, N. Genome-wide identification and expression analyses of cytochrome P450 genes in mulberry (Morus notabilis). J. Integr. Plant Biol. 2014, 56, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Racasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [PubMed]
- Cheng, J.; Kapranov, P.; Drenkow, J.; Dike, S.; Brubaker, S.; Patel, S.; Long, J.; Stern, D.; Tammana, H.; Helt, G. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005, 308, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.D.; Sung, S. Long noncoding RNA: Unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 2012, 17, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.H.; Wang, M.B. Molecular functions of long non-coding RNAs in plants. Genes 2012, 3, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Ann. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J.; Handley, P.; Wong, L.; McLachlan, D. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD). Neurochem. Res. 1992, 17, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Boerner, S.; McGinnis, K.M. Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS ONE 2012. [Google Scholar] [CrossRef] [PubMed]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Rymarquis, L.A.; Kastenmayer, J.P.; Hüttenhofer, A.G.; Green, P.J. Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 2008, 13, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Shuai, P.; Liang, D.; Tang, S.; Zhang, Z.; Ye, C.; Su, Y.; Xia, X.; Yin, W. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 2014. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Quan, M.; Zhang, D. Genome-wide identification of novel long non-coding RNAs in Populus tomentosa tension wood, opposite wood and normal wood xylem by RNA-seq. Planta 2015, 241, 125–143. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Qi, X.; Zeng, Q.; Xiang, Z.; He, N. MorusDB: A resource for mulberry genomics and genome biology. Database 2014. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005, 33, 501–504. [Google Scholar] [CrossRef] [PubMed]
- Flicek, P.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fizgerald, S. Ensembl 2014. Nucleic Acids Res. 2013. [Google Scholar] [CrossRef]
- Rosenbloom, K.R.; Armstrong, J.; Barber, G.P.; Casper, J.; Clawson, H.; Diekhans, M.; Dreszer, T.R.; Fujita, P.A.; Guruvadoo, L.; Haeussler, M. The UCSC genome browser database: 2015 Update. Nucleic Acids Res. 2015, 43, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Wilming, L.G.; Gilbert, J.G.R.; Howe, K.; Trevanion, S.; Hubbard, T.; Harrow, J.L. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008, 36, D753–D760. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Yuan, J.; Li, H.; Li, M.; Zhao, G.; Bu, D.; Zhu, W.; Wu, W.; Chen, R.; Zhao, Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014, 42, D98–D103. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.L.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.T.; Liao, Q.; Bu, D.; Zhao, G.; Liu, C.; Liu, Y.; Zhao, Y. Systematic study of human long intergenic non-coding RNAs and their impact on cancer. Sci. China Life Sci. 2013, 56, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Sun, L.; Li, P.; Bu, D.; Cao, H.; Zhao, Y. Comprehensive characterization of 10,571 mouse large intergenic noncoding RNAs from whole transcriptome sequencing. PLoS ONE. 2013, 8, e70835. [Google Scholar] [CrossRef] [PubMed]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploeta, L. The Arabidopsis information resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008, 36, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Liu, J.; Wang, H.; Wong, L.; Chua, N.H. PLncDB: Plant long noncoding RNA database. Bioinformatics 2013, 29, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myser, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.T.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Springer, P.S.; Holding, D.R.; Groover, A.; Yordan, C.; Martienssen, R.A. The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G (1) phase and is required maternally for early Arabidopsis development. Development 2000, 127, 1815–1822. [Google Scholar] [PubMed]
- Lin, Q.; Buckler, E.S.; Muse, S.V.; Walker, J.C. Molecular evolution of type 1 serine/threonine protein phosphatases. Mol. Phylogenet. Evolut. 1999, 12, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.D.; Walker, J.C. Plant protein phosphatases. Annu. Rev. Plant Biol. 1996, 47, 101–125. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Wang, M.; Wang, Q.; Wang, X.; Lin, J. Protein phosphatases 1 and 2A and the regulation of calcium uptake and pollen tube development in Picea wilsonii. Tree Physiol. 2006, 26, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Voll, L.; Häusler, R.E.; Hecker, R.; Weber, A.; Weissenböck, G.; Fiene, G.; Waffenschmidt, S.; Flügge, U.I. The phenotype of the Arabidopsis cue1 mutant is not simply caused by a general restriction of the shikimate pathway. Plant J. 2003, 36, 301–317. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tang, R.H.; Hao, Y.; Stevens, R.D.; Cook, C.W.; Ahn, S.M.; Jin, L.; Yang, Z.; Chen, L.; Guo, F. Nitric oxide represses the Arabidopsis floral transition. Science 2004, 305, 1968–1971. [Google Scholar] [CrossRef] [PubMed]
- Knappe, S.; Löttgert, T.; Schneider, A.; Voll, L.; Flügge, U.; Fischer, K. Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis–AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 2003, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, V.; Löttgert, T.; Geimer, S.; Dörmann, P.; Krüger, S.; Vijayakumar, V.; Schreiber, L.; Göbel, C.; Feussner, K.; Feussner, I. Phosphoenolpyruvate provision to plastids is essential for gametophyte and sporophyte development in Arabidopsis thaliana. Plant Cell 2010, 22, 2594–2617. [Google Scholar] [CrossRef] [PubMed]
- Shibagaki, N.; Rose, A.; McDermott, J.P.; Fujiwara, T.; Hayashi, H.; Yoneyama, T.; Davies, J.P. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1; 2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 2002, 29, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Buchner, P.; Parmar, S.; Kriegel, A.; Carpentier, M.; Hawkesford, M.J. The sulfate transporter family in wheat: Tissue-specific gene expression in relation to nutrition. Mol. Plant 2010, 3, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Krusell, L.; Krause, K.; Ott, T.; Desbrosses, T.; Krämer, U.; Sato, S.; Nakamura, Y.; Tabata, S.; James, E.K.; Sandal, N. The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 2005, 17, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Wang, Z.M.; Wang, M.; Wang, X.J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Wang, Y.; Yao, Y.; Song, N.; Hu, Z.; Qin, D.; Xie, C.; Peng, H.; Ni, Z.; Sun, Q. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, Q.; Carey, B.W.; Cassady, J.P. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Du, T.A. Non-coding RNA: RNA stability control by Pol II. Nat. Rev. Mol. Cell Biol. 2013, 14, 128–129. [Google Scholar]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Liao, Q.; Shen, J.; Liu, J.; Sun, X.; Zhao, G.; Chang, Y.; Xu, L.; Li, X.; Zhao, Y.; Zheng, H. Genome-wide identification and functional annotation of Plasmodium falciparum long noncoding RNAs from RNA-seq data. Parasitol. Res. 2014, 113, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Liao, J.Y.; Li, Z.Y.; Yu, Y.; Zhang, J.; Li, Q.; Qu, L.; Shu, W.; Chen, Y. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome biology 2014. [Google Scholar] [CrossRef] [PubMed]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Wahrisch, S.; Beisaw, A.; Macura, K.; Blass, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, N.; Yap, C.C.; Suzuki, M.; Kawai, K.; et al. Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566. [Google Scholar] [PubMed]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Pang, K.C.; Bruce, S.J.; Gardiner, B.B.; Askarian-Amiri, M.E.; Ru, K.; Soldà, G.; Simons, C. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008, 18, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. 2008, 105, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Li, J.H.; Jiang, S.; Zhou, H.; Qu, L.H. ChIPBase: A database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Rese. 2013, 41, D177–D187. [Google Scholar] [CrossRef] [PubMed]
- Sati, S.; Ghosh, S.; Jain, V.; Scaria, V.; Sengupta, S. Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic Acids Res. 2012, 40, 10018–10031. [Google Scholar] [CrossRef] [PubMed]



| Sample | Total Reads | Left Mapped Reads | Right Mapped Reads | Total Mapped Reads | |||
|---|---|---|---|---|---|---|---|
| Bark | 25,992,683 | 22,547,116 | 86.74% | 22,221,501 | 85.49% | 23,847,766 | 91.75% |
| Leaf | 24,809,215 | 22,686,967 | 91.45% | 22,244,419 | 89.66% | 24,123,695 | 97.24% |
| Root | 21,483,404 | 16,972,204 | 79.00% | 16,637,319 | 77.44% | 18,039,734 | 83.97% |
| Male flower | 26,629,083 | 24,015,382 | 90.18% | 23,545,895 | 88.42% | 25,681,360 | 96.44% |
| Winter bud | 18,138,525 | 14,706,155 | 81.08% | 14,392,578 | 79.35% | 15,841,259 | 87.33% |
| Sample | Junctions | Transcripts | Multi Exon | Multi Exon/Transcripts |
|---|---|---|---|---|
| Bark | 108814 | 30009 | 21907 | 73.00% |
| Leaf | 105808 | 32664 | 23354 | 71.50% |
| Root | 86084 | 28632 | 20163 | 70.42% |
| Male flower | 108894 | 35616 | 24368 | 68.42% |
| Winter bud | 75878 | 35553 | 21654 | 60.91% |
| Merge | 41042 | 30429 | 74.14% |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Sun, L.; Luo, H.; Ma, Q.; Zhao, Y.; Pei, D. Genome-Wide Identification and Characterization of Long Non-Coding RNAs from Mulberry (Morus notabilis) RNA-seq Data. Genes 2016, 7, 11. https://doi.org/10.3390/genes7030011
Song X, Sun L, Luo H, Ma Q, Zhao Y, Pei D. Genome-Wide Identification and Characterization of Long Non-Coding RNAs from Mulberry (Morus notabilis) RNA-seq Data. Genes. 2016; 7(3):11. https://doi.org/10.3390/genes7030011
Chicago/Turabian StyleSong, Xiaobo, Liang Sun, Haitao Luo, Qingguo Ma, Yi Zhao, and Dong Pei. 2016. "Genome-Wide Identification and Characterization of Long Non-Coding RNAs from Mulberry (Morus notabilis) RNA-seq Data" Genes 7, no. 3: 11. https://doi.org/10.3390/genes7030011





