Cloning and Expression Analysis of MEP Pathway Enzyme-encoding Genes in Osmanthus fragrans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Total RNA Extraction and Gene Cloning
2.3. Cloning of Full Length Genes by RACE
2.4. Gene Expression Analysis
3. Results
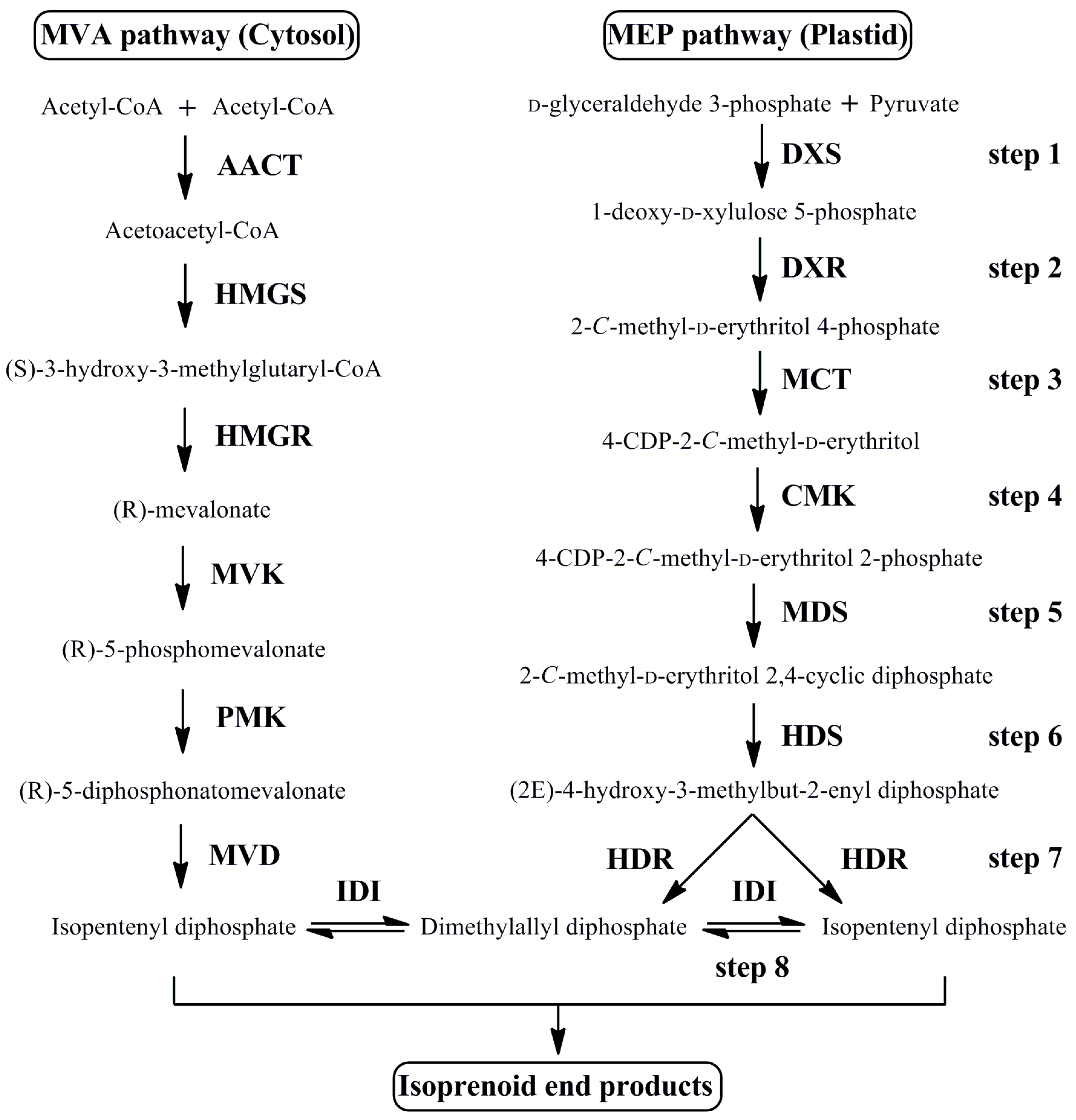
3.1. Cloning and Sequence Analysis of MEP Pathway Genes
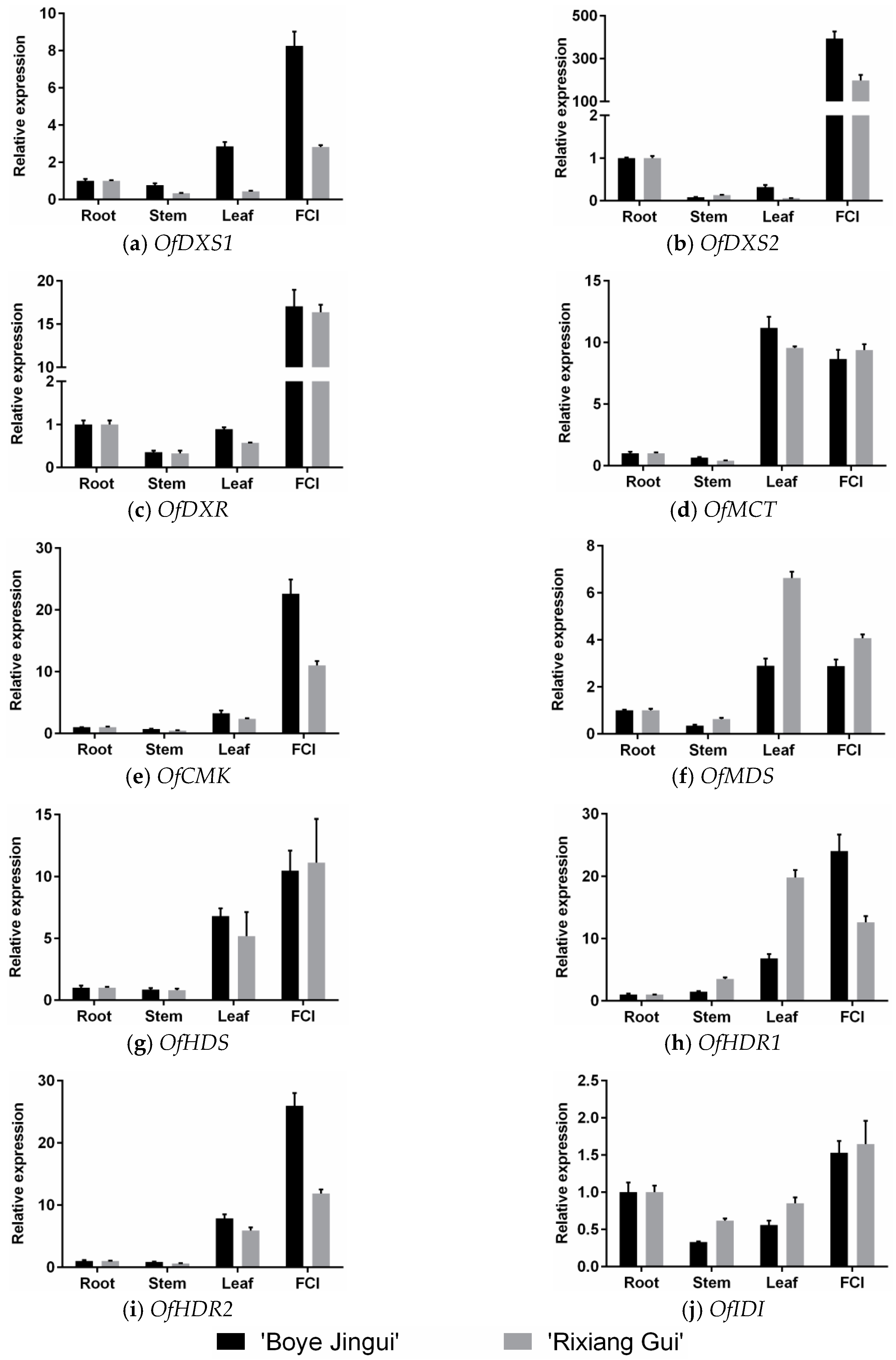
3.2. Expression Analysis of MEP Genes in Different Organs
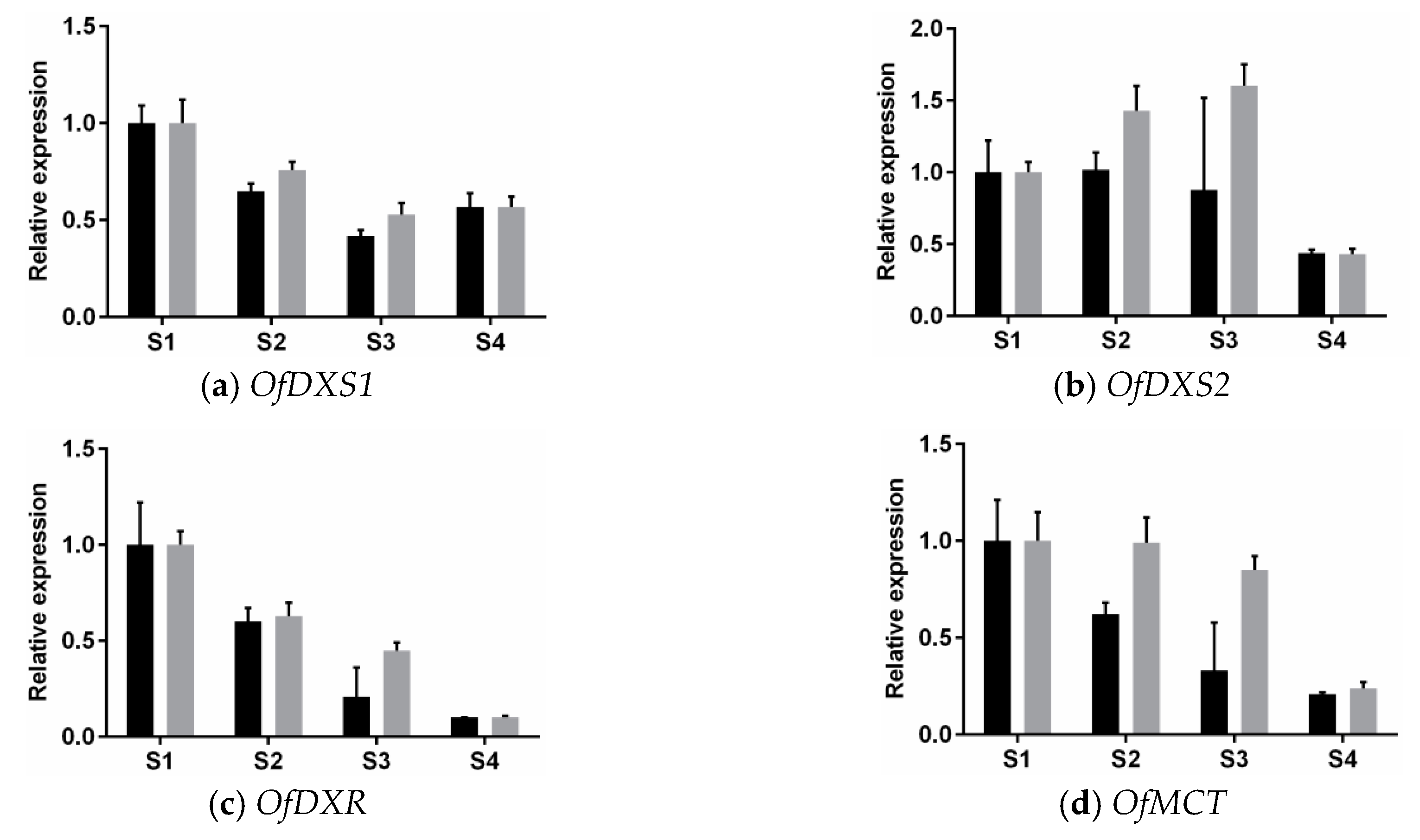
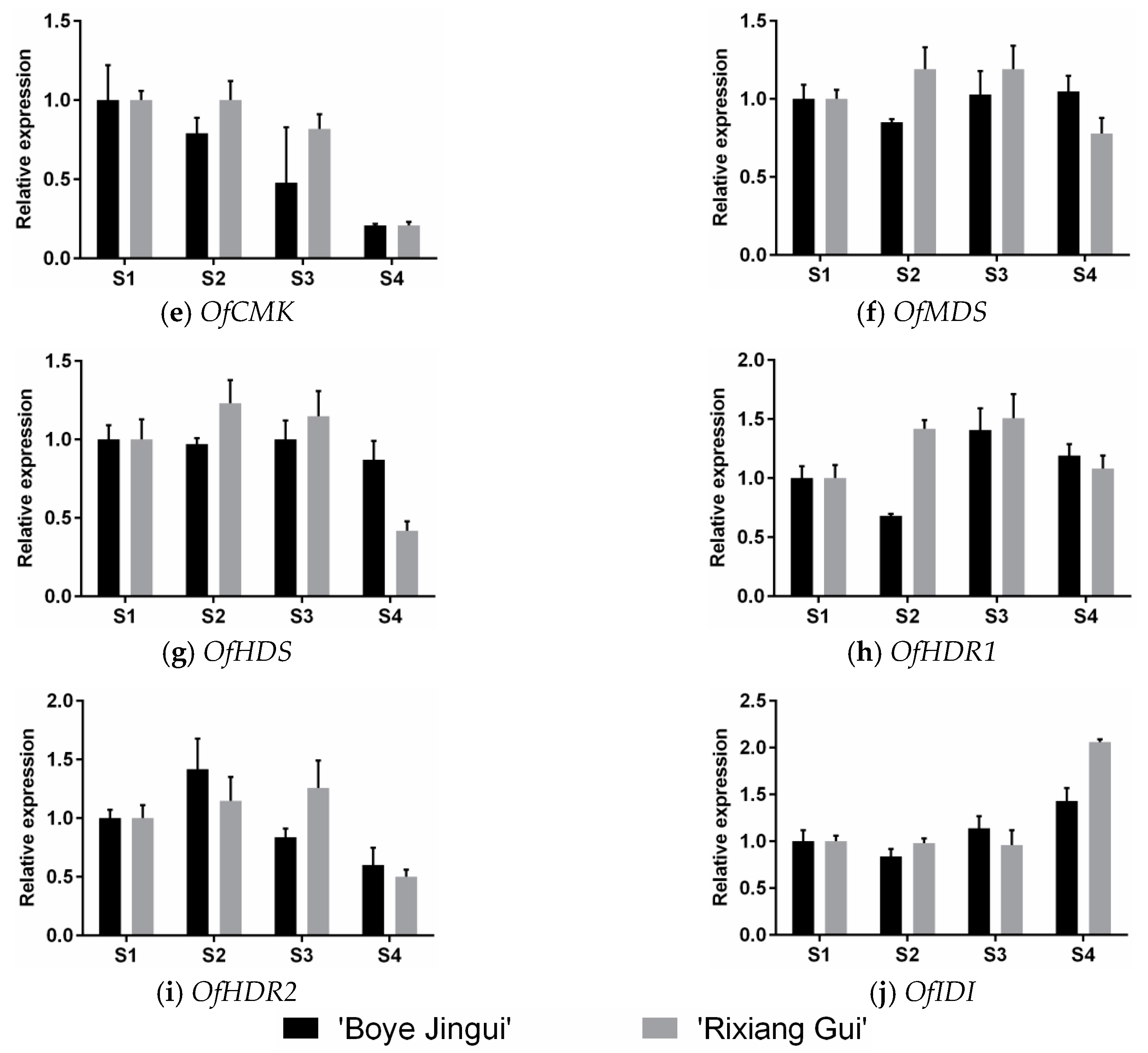
3.3. Expression Analysis of MEP Genes Over Flower Development
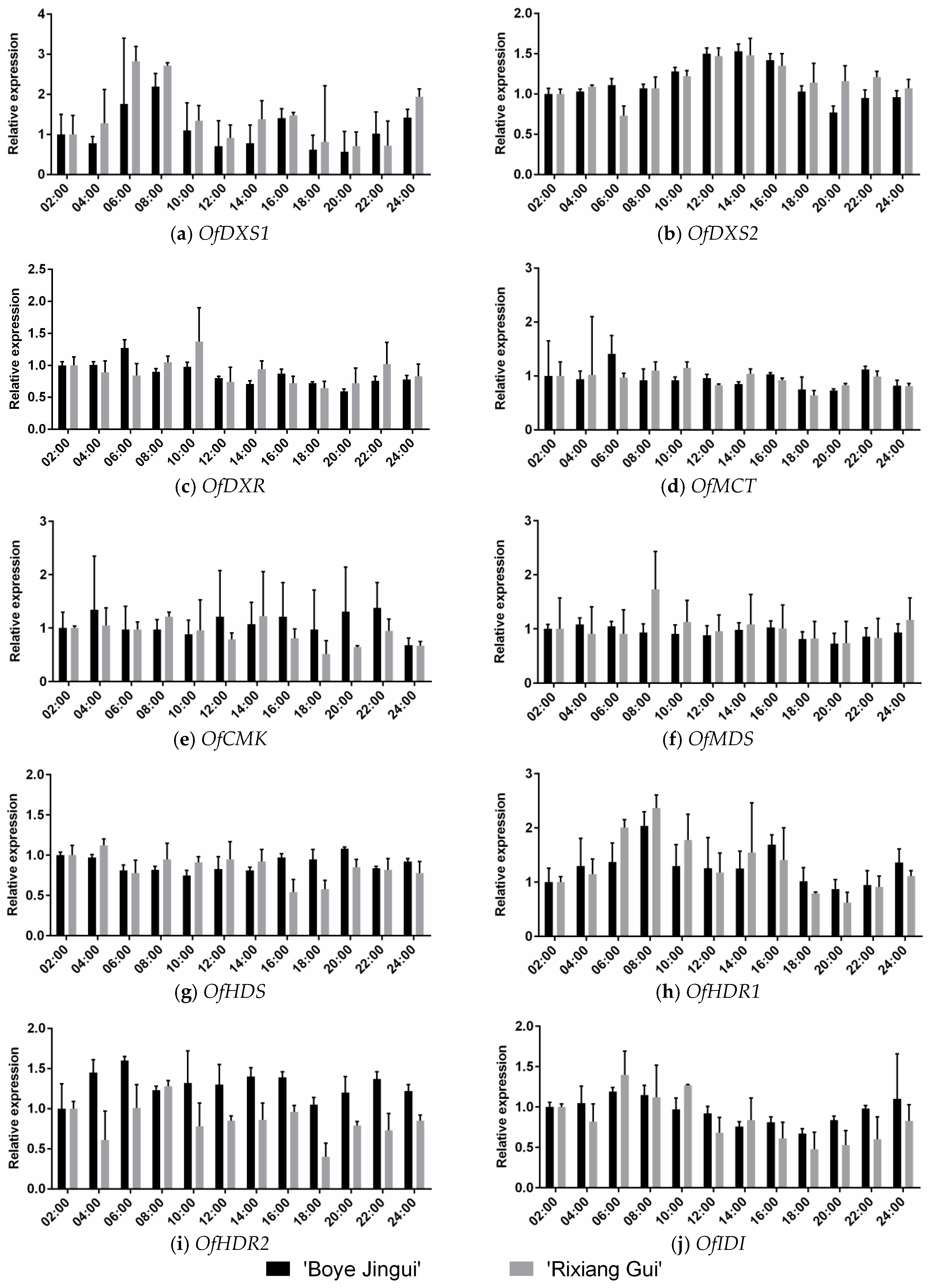
3.4. Expression analysis of MEP Genes during Diel Oscillations
4. Discussion
4.1. The MEP Pathway Genes of O. fragrans Are Highly Related to Those from Other Plants
4.2. Expression Patterns of the MEP Pathway Genes Suggest that Enhanced Biosynthesis of Substrate Contributes to the Production of Monoterpenes in O. fragrans Flowers
4.3. Expression Patterns of MEP Pathway Genes during Diel Oscillations
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DXS | 1-deoxy-d-xylulose-5-phosphate synthase |
| DXR | 1-deoxy-d-xylulose-5-phosphate reductoisomerase |
| MCT | 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase |
| CMK | 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase |
| MDS | 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase |
| HDS | 4-hydroxy-3-methylbut-2-enyl diphosphate synthase |
| HDR | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
| IDI | isopentenyl-diphosphate isomerase |
References
- Zhang, C.; Wang, Y.; Fu, J.; Bao, Z.; Zhao, H. Transcriptomic analysis and carotenogenic gene expression related to petal coloration in Osmanthus fragrans ‘Yanhong Gui’. Trees 2016, 1–17. [Google Scholar] [CrossRef]
- Baldermann, S.; Kato, M.; Kurosawa, M.; Kurobayashi, Y.; Fujita, A.; Fleischmann, P.; Watanabe, N. Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of Osmanthus fragrans Lour. J. Exp. Bot. 2010, 61, 2967–2977. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.B.; Liu, Y.L. Classification system of sweet osmanthus cultivars. In An Illustrated Monograph of the Sweet Osmanthus Cultivars in China; Zhejiang Science & Technology Press: Zhejiang, China, 2008; pp. 80–91. [Google Scholar]
- Yuan, W.J.; Li, Y.; Ma, Y.F.; Han, Y.J.; Shang, F.D. Isolation and characterization of microsatellite markers for Osmanthus fragrans (Oleaceae) using 454 sequencing technology. Genet. Mol. Res. 2015, 14, 4696–4702. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.J.; Chen, W.C.; Yang, F.B.; Wang, X.H.; Dong, M.F.; Zhou, P.; Shang, F.D. cDNA-AFLP analysis on 2 Osmanthus fragrans cultivars with different flower color and molecular characteristics of OfMYB1 gene. Trees 2015, 29, 931–940. [Google Scholar] [CrossRef]
- Deng, C.H.; Song, G.X.; Hu, Y.M. Application of HS-SPME and GC-MS to characterization of volatile compounds emitted from osmanthus flowers. Ann. Chim. 2004, 94, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Mai, R.Z.; Zou, J.J.; Zhang, H.Y.; Zeng, X.L.; Zheng, R.R.; Wang, C.Y. Analysis of aroma-active compounds in three sweet osmanthus (Osmanthus fragrans) cultivars by GC-olfactometry and GC-MS. J. Zhejiang Univ. B 2014, 15, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Baldermann, S.; Kato, M.; Fleischmann, P.; Watanabe, N. Biosynthesis of α- and β-ionone, prominent scent compounds, in flowers of osmanthus fragrans. Acta Biochim. Pol. 2012, 59, 79–81. [Google Scholar] [PubMed]
- Wang, L.M.; Li, M.T.; Jin, W.W.; Li, S.; Zhang, S.Q.; Yu, L.J. Variations in the components of Osmanthus fragrans Lour. essential oil at different stages of flowering. Food Chem. 2009, 114, 233–236. [Google Scholar] [CrossRef]
- Xin, H.P.; Wu, B.H.; Zhang, H.H.; Wang, C.Y.; Li, J.T.; Yang, B.; Li, S.H. Characterization of volatile compounds in flowers from four groups of sweet osmanthus (Osmanthus fragrans) cultivars. Can. J. Plant Sci. 2013, 93, 923–931. [Google Scholar] [CrossRef]
- Lei, G.M.; Mao, P.Z.; He, M.Q.; Wang, L.H.; Liu, X.S.; Zhang, A.Y. Water-soluble essential oil components of fresh flowers of Osmanthus fragrans Lour. J. Essent. Oil Res. 2016, 28, 177–184. [Google Scholar] [CrossRef]
- Zhang, C.; Fu, J.; Wang, Y.; Bao, Z.; Zhao, H. Identification of suitable reference genes for gene expression normalization in the quantitative real-time PCR analysis of sweet osmanthus (Osmanthus fragrans Lour.). PLoS ONE 2015, 10, e0136355. [Google Scholar] [CrossRef] [PubMed]
- Moronkola, D.O.; Aiyelaagbe, O.O.; Ekundayo, O. Syntheses of eight fragrant terpenoids [ionone derivatives] via the aldol condensation of citral and eight ketones. J. Essent. Oil Bear. Plants 2005, 8, 87–98. [Google Scholar] [CrossRef]
- Muhlemann, J.K.; Klempien, A.; Dudareva, N. Floral volatiles: From biosynthesis to function. Plant Cell Environ. 2014, 37, 1936–1949. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.L.; Liu, C.; Zheng, R.R.; Cai, X.; Luo, J.; Zou, J.J.; Wang, C.Y. Emission and accumulation of monoterpene and the key terpene synthase (TPS) associated with monoterpene biosynthesis in Osmanthus fragrans Lour. Front. Plant Sci. 2016, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. The 1-deoxy-d-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Gutensohn, M.; Orlova, I.; Nguyen, T.T.H.; Davidovich-Rikanati, R.; Ferruzzi, M.G.; Sitrit, Y.; Lewinsohn, E.; Pichersky, E.; Dudareva, N. Cytosolic monoterpene biosynthesis is supported by plastid-generated geranyl diphosphate substrate in transgenic tomato fruits. Plant J. 2013, 75, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Gahlan, P.; Kumar, S. Cloning and expression analysis of 10 genes associated with picrosides biosynthesis in Picrorhiza kurrooa. Gene 2013, 515, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Pulido, P.; Perello, C.; Rodríguez-Concepción, M. New insights into plant isoprenoid metabolism. Mol. Plant 2012, 5, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H. Non-mevalonate isoprenoid biosynthesis: Enzymes, genes and inhibitors. Biochem. Soc. Trans. 2000, 785–789. [Google Scholar] [CrossRef]
- Rohdich, F.; Kis, K.; Bacher, A.; Eisenreich, W. The non-mevalonate pathway of isoprenoids: Genes, enzymes and intermediates. Curr. Opin. Chem. Biol. 2001, 5, 535–540. [Google Scholar] [CrossRef]
- Hunter, W.N. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2007, 282, 21573–21577. [Google Scholar] [CrossRef] [PubMed]
- Cordoba, E.; Salmi, M.; León, P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 2009, 60, 2933–2943. [Google Scholar] [CrossRef] [PubMed]
- Rohmer, M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 1999, 16, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Estévez, J.M.; Cantero, A.; Romero, C.; Kawaide, H.; Jiménez, L.F.; Kuzuyama, T.; Seto, H.; Kamiya, Y.; León, P. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-d-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 2000, 124, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Carretero-paulet, L.; Ahumada, I.; Cunillera, N.; Rodrı́guez-Concepción, M.; Ferrer, A.; Boronat, A.; Campos, N. Expression and molecular analysis of the Arabidopsis DXR Gene encoding 1-deoxy-d-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-d-erythritol 4-phosphate pathway. Plant Physiol. 2002, 129, 1581–1591. [Google Scholar] [CrossRef] [PubMed]
- Chahed, K.; Oudin, A.; Guivarc, H.N.; Hamdi, S.; Chénieux, J.C.; Rideau, M.; Clastre, M. 1-deoxy-d-xylulose 5-phosphate synthase from periwinkle: cDNA identification and induced gene expression in terpenoid indole alkaloid-producing cells. Plant Physiol. Biochem. 2000, 38, 559–566. [Google Scholar] [CrossRef]
- Veau, B.; Courtois, M.; Oudin, A.; Chénieux, J.C.; Rideau, M.; Clastre, M. Cloning and expression of cDNAs encoding two enzymes of the MEP pathway in Catharanthus roseus. Biochim. Biophys. Acta 2000, 1517, 159–163. [Google Scholar] [CrossRef]
- Lange, B.M.; Wildung, M.R.; McCaskill, D.; Croteau, R. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 2100–2104. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.M.; Croteau, R. Isoprenoid biosynthesis via a mevalonate-independent pathway in plants: Cloning and heterologous expression of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from peppermint. Arch. Biochem. Biophys. 1999, 365, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Lois, L.M.; Rodríguez-Concepción, M.; Gallego, F.; Campos, N.; Boronat, A. Carotenoid biosynthesis during tomato fruit development: Regulatory role of 1-deoxy-d-xylulose 5-phosphate synthase. Plant J. 2000, 22, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Rohdich, F.; Wungsintaweekul, J.; Lüttgen, H.; Fischer, M.; Eisenreich, W.; Schuhr, C.A.; Fellermeier, M.; Schramek, N.; Zenk, M.H.; Bacher, A. Biosynthesis of terpenoids: 4-diphosphocytidyl-2-C- methyl-d-erythritol kinase from tomato. Proc. Natl. Acad. Sci. USA 2000, 97, 8251–8256. [Google Scholar] [CrossRef] [PubMed]
- Cordoba, E.; Porta, H.; Arroyo, A.; San Román, C.; Medina, L.; Rodríguez-Concepción, M.; León, P. Functional characterization of the three genes encoding 1-deoxy-d-xylulose 5-phosphate synthase in maize. J. Exp. Bot. 2011, 62, 2023–2038. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Kuzuyama, T.; Kobayashi, A.; Sando, T.; Chang, Y.J.; Kim, S.U. 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase (IDS) is encoded by multicopy genes in gymnosperms Ginkgo biloba and Pinus taeda. Planta 2008, 227, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.R.; Su, P.; Zhao, Y.J.; Zhang, M.; Wang, X.J.; Liu, Y.J.; Zhang, X.N.; Gao, W.; Huang, L.Q. Molecular cloning and characterization of DXS and DXR genes in the terpenoid biosynthetic pathway of Tripterygium wilfordii. Int. J. Mol. Sci. 2015, 16, 25516–25535. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Zeng, L.; Yuan, Y.; Chen, M.; Wang, F.; Liu, X.; Zeng, L.; Lan, X.; Liao, Z. Enhancement of artemisinin biosynthesis by overexpressing dxr, cyp71av1 and cpr in the plants of Artemisia annua L. Plant Omics 2012, 5, 503–507. [Google Scholar]
- Roberts, S.C. Production and engineering of terpenoids in plant cell culture. Nat. Chem. Biol. 2007, 3, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.N.; Li, H.G.; Wang, L.G.; Yang, X.L.; Sun, T.Z.; Xu, C. Transcriptome sequencing and analysis of sweet osmanthus (Osmanthus fragrans Lour.). Genes Genomics 2014, 36, 777–788. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.M.; Zhang, L.; Wang, J.; Liao, P.; Zhang, Y.; Zhang, R.; Kai, G.Y. Molecular characterization and expression of 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR) gene from Salvia miltiorrhiza. Acta Physiol. Plant 2009, 31, 1015–1022. [Google Scholar] [CrossRef]
- Munos, J.W.; Pu, X.; Mansoorabadi, S.O.; Kim, H.J.; Liu, H.W. A secondary kinetic isotope effect study of the 1-deoxy-d-xylulose-5-phosphate reductoisomerase-catalyzed reaction: Evidence for a retroaldol-aldol rearrangement. J. Am. Chem. Soc. 2009, 131, 2048–2049. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Paulet, L.; Cairó, A.; Botella-Pavía, P.; Besumbes, O.; Campos, N.; Boronat, A.; Rodríguez-Concepción, M. Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol. Biol. 2006, 62, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Rohdich, F.; Wungsintaweekul, J.; Eisenreich, W.; Richter, G.; Schuhr, C.A.; Hecht, S.; Zenk, M.H.; Bacher, A. Biosynthesis of terpenoids: 4-diphosphocytidyl-2C-methyl-d-erythritol synthase of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 6451–6456. [Google Scholar] [CrossRef] [PubMed]
- Dubey, V.S.; Bhalla, R.; Luthra, R. An overview of the non-mevalonate pathway for terpenoid biosynthesis in plants. J. Biosci. 2003, 28, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Kuzuyama, T.; Chang, Y.J.; Kim, S.U. Cloning and characterization of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase (MECS) gene from Ginkgo biloba. Plant Cell Rep. 2006, 25, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Sedkova, N.; Tao, L.; Rouvière, P.E.; Cheng, Q. Diversity of carotenoid synthesis gene clusters from environmental enterobacteriaceae strains. Appl. Environ. Microbiol. 2005, 71, 8141–8146. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Cseke, L.; Blanc, V.M.; Pichersky, E. Evolution of floral scent in Clarkia: Novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 1996, 8, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.A.; Gutensohn, M.; Wilkerson, C.G.; Dudareva, N. Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 2008, 55, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tholl, D.; D’Auria, J.C.; Farooq, A.; Pichersky, E.; Gershenzon, J. Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 2003, 15, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Dudareva, N.; Bhakta, S.; Raguso, R.A.; Pichersky, E. Floral scent production in Clarkia breweri. Plant Physiol. 1998, 116, 599–604. [Google Scholar]
- Xiang, L.; Zhao, K.G.; Chen, L.Q. Molecular cloning and expression of Chimonanthus praecox farnesyl pyrophosphate synthase gene and its possible involvement in the biosynthesis of floral volatile sesquiterpenoids. Plant Physiol. Biochem. 2010, 48, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.J.; Wang, X.H.; Chen, W.C.; Dong, M.F.; Yuan, W.J.; Liu, X.; Shang, F.D. Differential expression of carotenoid-related genes determines diversified carotenoid coloration in flower petal of Osmanthus fragrans. Tree Genet. Genomes 2014, 10, 329–338. [Google Scholar] [CrossRef]
- Yang, X.L.; Shi, T.T.; Wen, A.L.; Wang, L.G. Variance analysis of aromatic components from different varieties of Osmanthus fragrans. Dongbei Linye Daxue Xuebao 2015, 43, 83–87. (In Chinese) [Google Scholar]
- Nagegowda, D.A.; Rhodes, D.; Dudareva, N. The role of methylerythritol-phosphate pathway in rhythmic emission of volatiles. In Biology of Floral Scent; CRC Press: Boca Raton, FL, USA, 2006; pp. 139–153. [Google Scholar]
- Mandel, M.A.; Feldmann, K.A.; Herrera-Estrella, L.; Rocha-Sosa, M.; Leon, P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996, 9, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Andersson, S.; Orlova, I.; Gatto, N.; Reichelt, M.; Rhodes, D.; Boland, W.; Gershenzon, J. The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 2005, 102, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Hendel-Rahmanim, K.; Masci, T.; Vainstein, A.; Weiss, D. Diurnal regulation of scent emission in rose flowers. Planta 2007, 226, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Pokhilko, A.; Bou-Torrent, J.; Pulido, P.; Rodríguez-Concepción, M.; Ebenhöh, O. Mathematical modelling of the diurnal regulation of the MEP pathway in Arabidopsis. New Phytol. 2015, 206, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
| Gene | Accession No. | Full Length (bp) | ORF (bp) | Amino Acids (aa) | Molecular Weight (kDa) | PI |
|---|---|---|---|---|---|---|
| OfDXS1 | KX400841 | 2645 | 2172 | 723 | 78.1 | 6.91 |
| OfDXS2 | KX400842 | 2533 | 2148 | 715 | 76.9 | 6.91 |
| OfDXR | KX400843 | 1687 | 1425 | 474 | 51.3 | 6.04 |
| OfMCT | KX400844 | 1155 | 939 | 312 | 34.4 | 7.67 |
| OfCMK | KX400845 | 1543 | 1206 | 401 | 44.4 | 5.75 |
| OfMDS | KX400846 | 991 | 702 | 233 | 25.1 | 8.64 |
| OfHDS | KX400847 | 2551 | 2229 | 742 | 82.5 | 5.78 |
| OfHDR1 | KX400848 | 1642 | 1386 | 461 | 52.0 | 5.51 |
| OfHDR2 | KX400849 | 1593 | 1380 | 459 | 51.8 | 5.73 |
| OfIDI | KX400850 | 1130 | 708 | 235 | 26.9 | 5.14 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Li, H.; Yang, X.; Gu, C.; Mu, H.; Yue, Y.; Wang, L. Cloning and Expression Analysis of MEP Pathway Enzyme-encoding Genes in Osmanthus fragrans. Genes 2016, 7, 78. https://doi.org/10.3390/genes7100078
Xu C, Li H, Yang X, Gu C, Mu H, Yue Y, Wang L. Cloning and Expression Analysis of MEP Pathway Enzyme-encoding Genes in Osmanthus fragrans. Genes. 2016; 7(10):78. https://doi.org/10.3390/genes7100078
Chicago/Turabian StyleXu, Chen, Huogen Li, Xiulian Yang, Chunsun Gu, Hongna Mu, Yuanzheng Yue, and Lianggui Wang. 2016. "Cloning and Expression Analysis of MEP Pathway Enzyme-encoding Genes in Osmanthus fragrans" Genes 7, no. 10: 78. https://doi.org/10.3390/genes7100078





