Genomic Distribution and Divergence of Levansucrase-Coding Genes in Pseudomonas syringae
Abstract
:1. Introduction
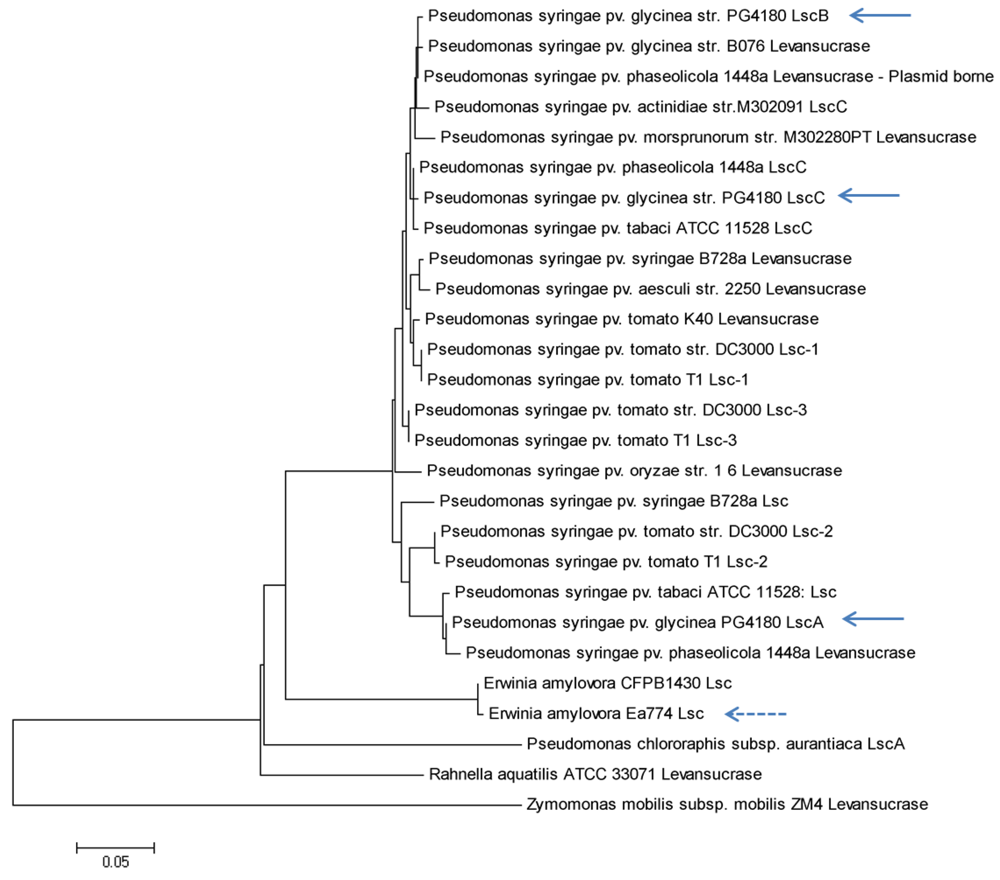
2. Results and Discussion
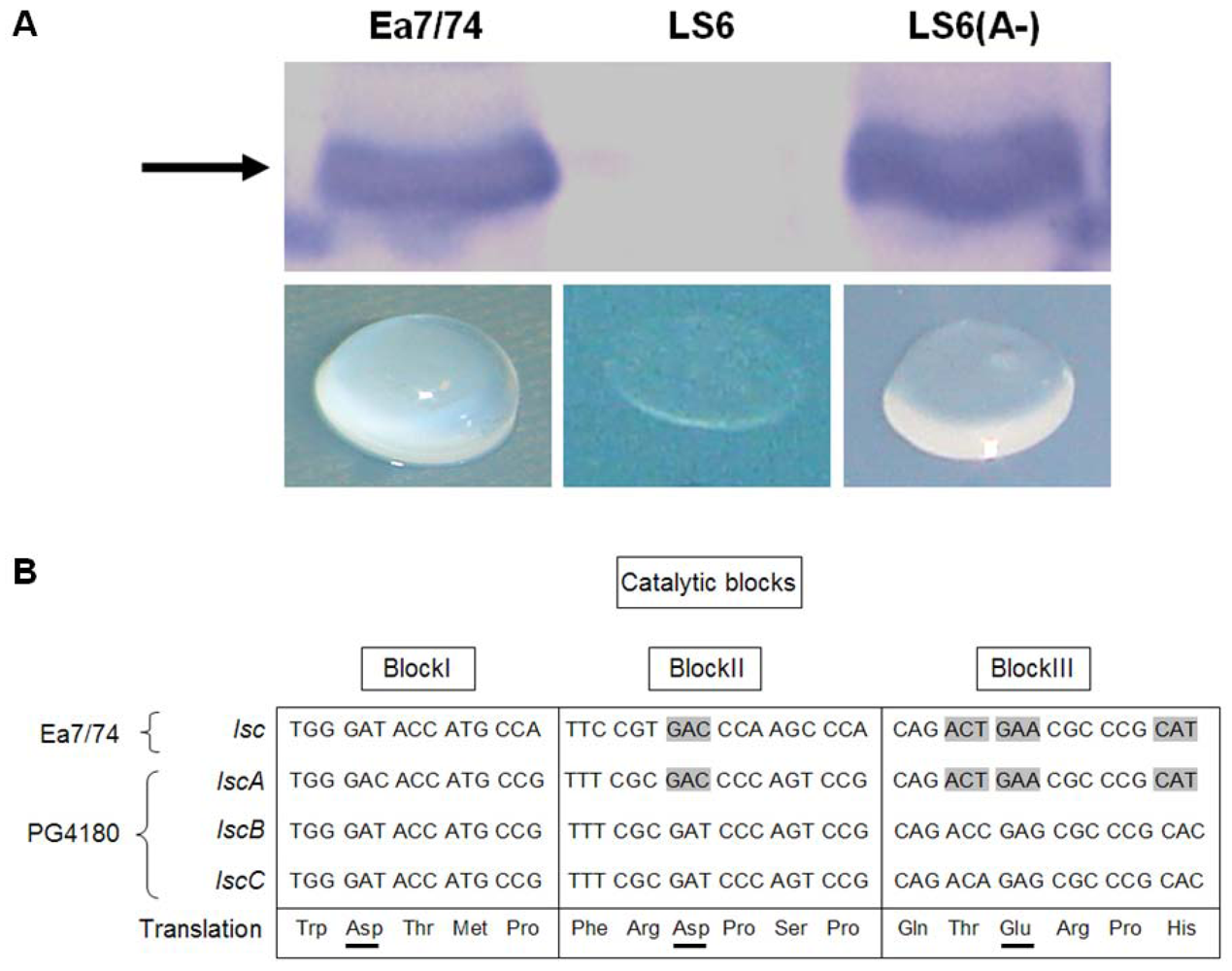
2.1. Heterologous Expression of lscA in E. amylovora
2.2. Nucleotide Sequence Comparison of Variant A lsc Genes
| variant A (1,248 bp) | variant BC (1,296 bp) | |||||||
| Pathovar, strain | Gene name | Locus tag | Genomic location | Gene name | Locus tag | Genomic location | References | |
| Pseudomonadaceae | ||||||||
| Pseudomonas syringae | glycinea PG4180 | lscA | - | Chr. | lscB | - | Plasmid | [3] |
| lscC | - | Chr. | ||||||
| glycinea B076 | levansucrase | PsgB076_10300 | ? | lscC | PsgB076_00457 | ? | [9] | |
| glycinea race 4 | levansucrase | PsgRace4_15609 | ? | lscC | PsgRace4_03819 | ? | [9] | |
| phaseolicola 1448A | levansucrase | PSPPH_2074 | Chr. | levansucrase | PSPPH_A0027 | Plasmid | [6] | |
| lscC | PSPPH_4994 | Chr. | ||||||
| syringae B728a | levansucrase | Psyr_2103 | Chr. | levansucrase | Psyr_0754 | Chr. | [7] | |
| actinidiae M302091 | - | lscC | PSYAC_19498 | ? | [42] | |||
| aesculi NCPPB3681 | levansucrase | PsyrpaN_010100019209 | ? | - | [43] | |||
| lachrymans M302278PT | levansucrase | PLA107_25445 | ? | - | [43] | |||
| morsprunorum M302280PT | - | levansucrase | PSYMP_24576 | ? | [43] | |||
| tabaci ATCC 11528 | levansucrase | PSYTB_12850 | ? | lscC | PsyrptA_020100005135 | ? | [42,43] | |
| tomato DC3000 | lsc-2 | PSPTO_2305 | Chr. | lsc-3 | PSPTO_A0032 | Plasmid | [5] | |
| lsc-1 | PSPTO_1453 | Chr. | ||||||
| tomato T1 | lsc-2 | PSPTOT1_4965 | Chr. | lsc-3 | PSPTOT1_4913 | ? | [8] | |
| lsc-1 | PSPTOT1_1070 | Chr. | ||||||
| tomato K40 | - | levansucrase | PsyrptK_010100027584 | ? | Vinatzer et al. (unpublished) Genbank | |||
| tomato NCPPB 1108 | levansucrase | PsyrptN_010100027628 | ? | - | Vinatzer et al. (ubpublished) Genbank | |||
| Enterobacteriaceae | ||||||||
| Erwinia amylovora | Ea7/74 | Lsc | - | Chr. | - | [15] | ||
| CFPB1430 | Lsc | - | Chr. | - | [44] | |||
| Erwinia tasmaniensis | Et1/99 | Lsc | ETA_34670 | Chr. | - | [45] | ||
| Rahnella aquatilis | ATCC33071 | lsrA | - | Chr. | - | [46] | ||
2.3. Nucleotide Sequence Comparison of Variant BC lsc Genes

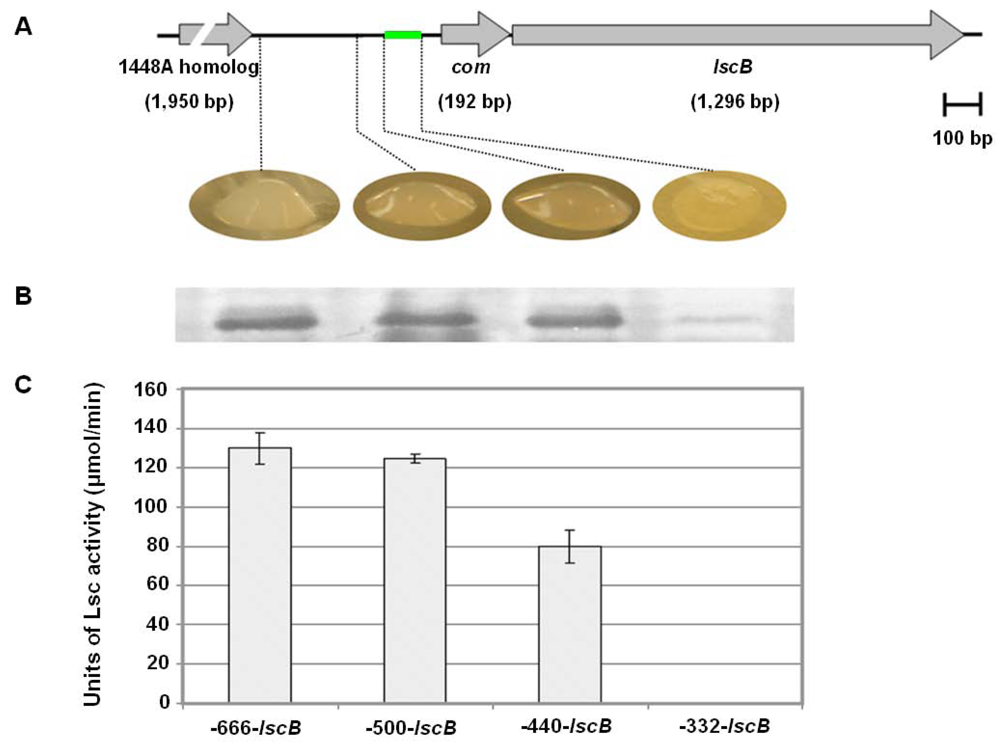
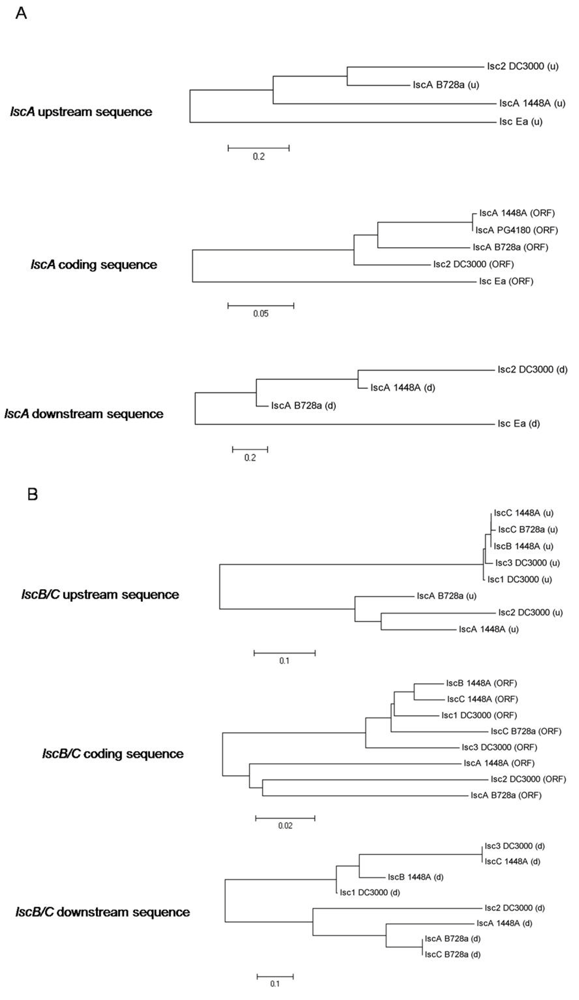
2.4. Downstream Sequence Comparison of Variant BC lsc Alleles
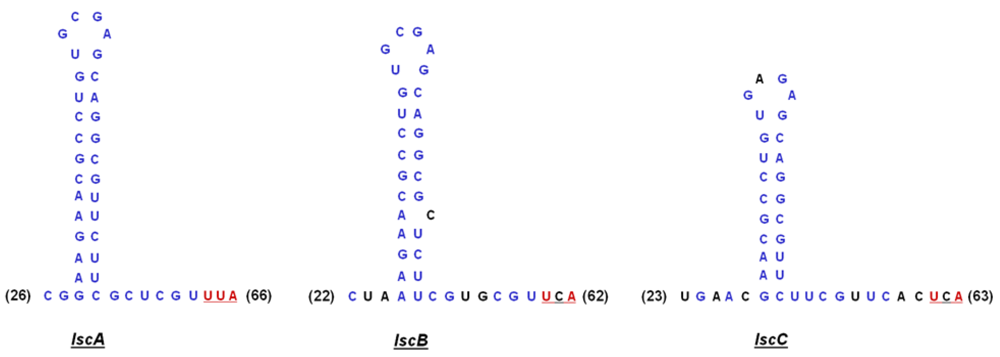
2.5. Upstream Sequence Comparison of Variant BC lsc Alleles and a Putative Prophage-Borne Glycosyl Hydrolase Gene
| PAPE association with | Genomic location | Length (bp) |
|---|---|---|
| Prophage PSPPH01, putative cellulase (PSPPH 0655), 1448A chromosome #§ | 773003–773504 | 502 |
| putative GH5 Cellulase, (Psyr_4600), B728a chromosome #§ | 5460116–5459615 | 502 |
| putative bacteriocin, (PSPTO_0572), DC3000 chromosome #§ | 629397–629790 * | 394 |
| lsc-1/C, DC3000 chromosome # | 1595373–1594873 | 501 |
| lsc-3/B, DC3000 plasmid pDC3000A # | 34651–34152 | 500 |
| lscC (Psyr_0754), B728a chromosome # | 859840–859339 | 502 |
| lscB (PSPPH_A0027), 1448A large plasmid # | 22669–22170 | 500 |
| lscC (PSPPH_4994), 1448A chromosome # | 5662790–5663289 | 500 |
| lscB, PG4180, plasmid # | - | 500 |
| lscC, PG4180, chromosome # | - | 500 |
| glycosyl hydrolase, PG4180 ¤ | - | 502 |
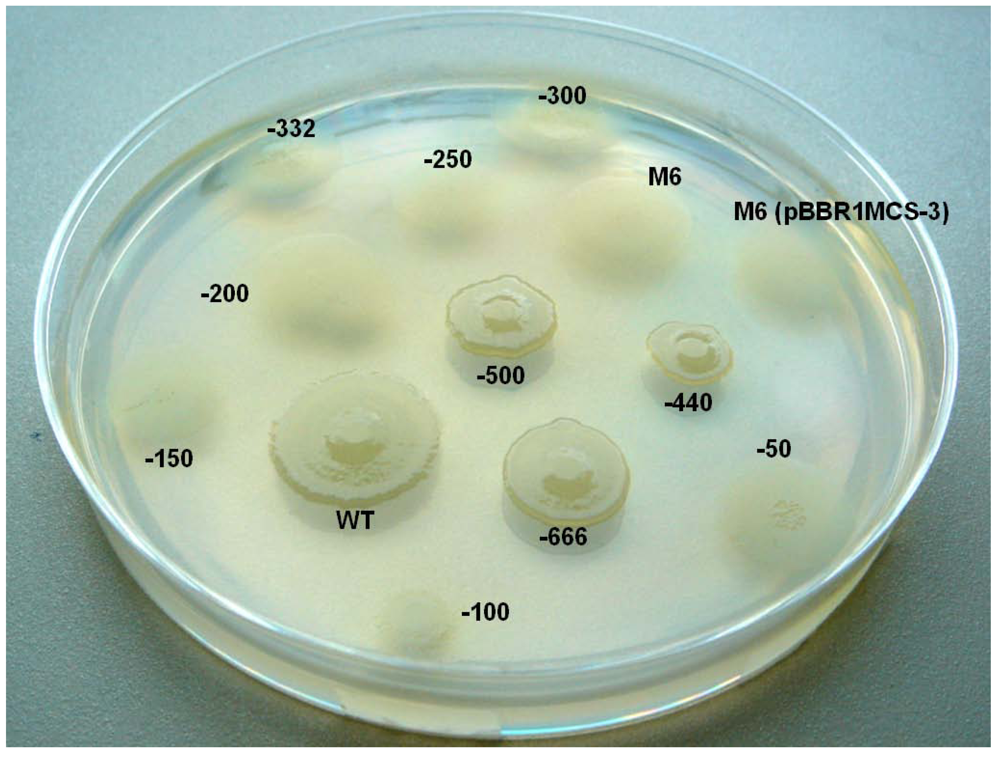
2.6. Investigating the Role of com Gene and Glycosyl Hydrolase Gene in PG4180
2.7. Putative Scenario for lsc Gene Distribution in P. syringae
3. Materials and Methods
3.1. Bacterial Strains, Plasmids and Growth Conditions
3.2. Molecular Genetic Techniques
| Strain | Relevant characteristicsa | Reference or source |
|---|---|---|
| Pseudomonas syringae pv. Glycinea | ||
| PG4180 | wild type, levan+ | [47] |
| PG4180.M6 | Spr, Gmr, lscB lscC mutant of PG4180, levan- | [3] |
| PG4180.M6 (pBBR1MCS-3) | Spr, Gmr, Tcr, lscB lscC mutant of PG4180 bearing pBBR1MCS | This Study |
| Erwinia amylovora | ||
| Ea7/74 | Ea7/74 | Ea7/74 |
| Ea7/74-LS6 | Ea7/74-LS6 | Ea7/74-LS6 |
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (Φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | [35] |
| pSKL3 | Apr, contains lscA on 3.0-kb Pst1 insert, (Plac > lscA) | [3] |
| pBBR3(lscA) | Tcr, contains lscA on 3.0-kb Pst1 insert, (lscA > Plac) | This study |
| pBBR1MCS | Cmr, broad-host-range cloning vector | [37] |
| pBBR1MCS-3 | Tcr, broad-host-range cloning vector | [37] |
| -666-lscB | Tcr, lscB gene with -666bp upstream sequence in pBBR1MCS-3 | This study |
| -500-lscB | Tcr, lscB gene with -500bp upstream sequence in pBBR1MCS-3 | This study |
| -440-lscB | Tcr, lscB gene with -440bp upstream sequence in pBBR1MCS-3 | This study |
| -332-lscB | Tcr, lscB gene with -332bp upstream sequence in pBBR1MCS-3 | This study |
| -300-lscB | Tcr, lscB gene with -300bp upstream sequence in pBBR1MCS-3 | This study |
| -250-lscB | Tcr, lscB gene with -250bp upstream sequence in pBBR1MCS-3 | This study |
| -200-lscB | Tcr, lscB gene with -200bp upstream sequence in pBBR1MCS-3 | This study |
| -150-lscB | Tcr, lscB gene with -150bp upstream sequence in pBBR1MCS-3 | This study |
| -100-lscB | Tcr, lscB gene with -100bp upstream sequence in pBBR1MCS-3 | This study |
| -50-lscB | Tcr, lscB gene with -50bp upstream sequence in pBBR1MCS-3 | This study |
| -666-lscC | Tcr, lscC gene with -327bp upstream sequence in pBBR1MCS-3 | This study |
| -500-lscC | Tcr, lscC gene with -161bp upstream sequence in pBBR1MCS-3 | This study |
| -440-lscC | Tcr, lscC gene with -101bp upstream sequence in pBBR1MCS-3 | This study |
| -332-lscC | Tcr, lscC gene with -332bp upstream sequence in pBBR1MCS-3 | This study |
| -666-lscB.com1 | Cmr, lscB gene with -666 upstream sequence in pBBR1MCS, com gene containing a premature stop codon | This study |
| Oligonucleotides | Nucleotide sequence (5’-3’) a |
|---|---|
| lscB_PG-666_fwd | GATGAGCTCCTAAGGCAGTCGCATTAA |
| lscB_PG-500_fwd | GATGAGCTCAGTCGCAATTAATGCGAG |
| lscB_PG-440_fwd | GATGAGCTCCCAGGTCAATGGCGCAGC |
| lscB_PG-332_fwd | GATGAGCTCCACGATATGCGATTTGCG |
| lscB_PG-300_fwd | GATGAGCTCCCGGATACGGGCTTTTAA |
| lscB_PG-250_fwd | GATGAGCTCACCCCGCCCAGCCGGGGT |
| lscB_PG-200_fwd | GATGAGCTCCAAATGTTGAAAGACTAC |
| lscB_PG-150_fwd | GATGAGCTCCATGGGTGACTACACCGA |
| lscB_PG-100_fwd | GATGAGCTCTGAATCATGTGAAGGCCG |
| lscB_PG-50_fwd | GATGAGCTCGGTACACGAGCGTCGCTG |
| lscB_PG_rev | CGATCTAGATCAGCTTAGCGTCACGTC |
| lscC_PG-666_fwd | GATGAGCTCAGCTCTGCCAGAAACAGG |
| lscC_PG-500_fwd | GATGAGCTCTCATAGGAAATTCCTTTT |
| lscC_PG-440_fwd | GATGAGCTCCCGGGTCAATTGCGCAAC |
| lscC_PG-332_fwd | GATGAGCTCCACGATATGCGATTTGCG |
| lscC_PG_rev | CGATCTAGATCAGCTCAGTTGCACGTC |
| com1 | GCAAATGTTGAAAGACTACCGATGCGGGCAGTGC |
| lscBC_RT_fwd | TCGGTTATCCTGACCCTGAC |
| lscBC_RT_rev | CCATGACGATCTTCCCAGTC |
| cel_RT_fwd | ACAAGATGGCCGCTTTATC |
| cel_RT_rev | TTCGCTTTATCGAGCAGGTT |
3.3. Extra-Cellular Lsc Detection
3.4. Analysis of Glycosyl Hydrolase gene Expression by Quantitative Reverse-Transcriptase Polymerase Chain Reaction (qRT-PCR)
3.5. Bioinformatics Analyses
4. Conclusions and Future Scope
Acknowledgments
References
- Young, J.M.; Saddler, G.S.; Takikawa, Y.; De Boer, S.H.; Vauterin, L.; Gardan, L.; Gvozdyak, R.I.; Stead, D.E. Names of plant pathogenic bacteria 1864–1995. Rev. Plant Pathol. 1996, 75, 721–763. [Google Scholar]
- Hettwer, U.; Jaeckel, F.R.; Boch, J.; Meyer, M.; Rudolph, K.; Ullrich, M.S. Cloning, nucleotide sequence, and expression in Escherichia coli of levansucrase genes from the plant pathogens Pseudomonas syringae pv.glycinea and P. syringae pv. phaseolicola. Appl. Environ. Microbiol. 1998, 64, 3180–3187. [Google Scholar]
- Li, H.; Ullrich, M.S. Characterization and mutational analysis of three allelic lsc genes encoding levansucrase in Pseudomonas syringae. J. Bacteriol. 2001, 183, 3282–3292. [Google Scholar] [CrossRef]
- Li, H.; Schenk, A.; Srivastava, A.; Zhurina, D.; Ullrich, M.S. Thermoresponsive expression and differential secretion of the extracellular enzyme levansucrase in the plant pathogenic bacterium Pseudomonas syringae. FEMS Microbiol. Lett. 2006, 265, 178–185. [Google Scholar] [CrossRef]
- Buell, C.R.; Joardar, V.; Lindeberg, M.; Selengut, J.; Paulsen, I.T.; Gwinn, M.L.; Dodson, R.J.; Deboy, R.T.; Durkin, A.S.; Kolonay, J.F.; et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2003, 100, 10181–10186. [Google Scholar]
- Joardar, V.; Lindeberg, M.; Jackson, R.W.; Selengut, J.; Dodson, R.; Brinkac, L.M.; Daugherty, S.C.; Deboy, R.; Durkin, A.S.; Giglio, M.G. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 2005, 187, 6488–6498. [Google Scholar]
- Feil, H.; Feil, W.S.; Chain, P.; Larimer, F.; Di Bartolo, G.; Copeland, A.; Lykidis, A.; Trong, S.; Nolan, M.; Goltsman, E.; et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA. 2005, 102, 11064–11069. [Google Scholar]
- Almeida, N.F.; Yan, S.; Lindeberg, M.; Studholme, D.J.; Schneider, D.J.; Condon, B.; Liu, H.; Viana, C.J.; Warren, A.; Evans, C; et al. A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a type III effector repertoire significantly divergent from that of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 2009, 22, 52–62. [Google Scholar] [CrossRef]
- Qi, M.; Wang, D.; Bradley, C.A.; Zhao, Y. Genome sequence analyses of Pseudomonas savastanoi pv. glycinea and subtractive hybridization-based comparative genomics with nine pseudomonads. PLoS One 2011, 6. [Google Scholar]
- Visnapuu, T.; Mäe, A.; Alamäe, T. Hansenula polymorpha maltase gene promoter with sigma 70-like elements is feasible for Escherichia coli-based biotechnological applications: Expression of three genomic levansucrase genes of Pseudomonas syringae pv. tomato. Process Biochem. 2008, 43, 414–422. [Google Scholar] [CrossRef]
- Visnapuu, T.; Zamfir, A.D.; Mosoarca, C.; Stanescu, M.D.; Alamäe, T. Fully automated chip-based negative mode nanoelectrospray mass spectrometry of fructooligosaccharides produced by heterologously expressed levansucrase from Pseudomonas syringae pv. tomato DC3000. Rapid Commun. Mass Spec. 2009, 23, 1337–1346. [Google Scholar] [CrossRef]
- Visnapuu, T.; Mardo, K.; Mosoarca, C.; Zamfir, A.D.; Vigants, A.; Alamäe, T. Levansucrases from Pseudomonas syringae pv. tomato and P. chlororaphis subsp. aurantiaca: Substrate specificity, polymerizing properties and usage of different acceptors for fructosylation. J. Biotechnol. 2011, 155, 34–39. [Google Scholar]
- Sarkar, S.F.; Guttman, D.S. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 2004, 70, 1999–2012. [Google Scholar]
- Falkenstein, H.; Bellemann, P.; Walter, S.; Zeller, W.; Geider, K. Identification of Erwinia amylovora, the fireblight pathogen, by colony hybridization with DNA from plasmid pEA29. Appl. Environ. Microbiol. 1988, 54, 2798–2802. [Google Scholar]
- Geier, G.; Geider, K. Characterization and influence on virulence of the levansucrase gene from the fire blight pathogen Erwinia amylora. Physiol. Mol. Plant Pathol. 1993, 42, 387–404. [Google Scholar] [CrossRef]
- Dols, M.; Remaud-Simon, M.; Villemot, R.M.; Vignon, V.; Mosnsan, P. Characterization on the different dextransucrases excreted in glucose, fructose or sucrose medium by Leuconostoc mesenteroides B 1299. Appl. Environ. Microbiol. 1998, 64, 1298–1302. [Google Scholar]
- Kim, K.Y.; Jordan, D.; Krishnan, H.B. Rahnella aquatilis, a bacterium isolated from soybean rhizosphere, can solubilize hydroxyapatite. FEMS Microbiol. Lett. 1997, 153, 273–277. [Google Scholar] [CrossRef]
- Pons, T.; Naumoff, D.G.; Martínez-Fleites, C.; Hernández, L. Three acidic residues are at the active site of a β-propeller architecture in glycoside hydrolase families 32, 43, 62, and 68. Proteins 2004, 54, 424–432. [Google Scholar]
- Lammens, W.; LeRoy, K.; Schroeven, L.; Van Laere, A.; Rabijns, A.; van den Ende, W. Structural insights into glycoside hydrolase family 32 and 68 enzymes: Functional implications. J. Exp. Bot. 2009, 60, 727–740. [Google Scholar]
- Sesma, A.; Murillo, J. The biochemistry and molecular genetics of host range definition: Pseudomonas syringae. Phytopathol. Mediterr. 2001, 40, 3–26. [Google Scholar]
- Polard, P.; Prère, M.F.; Fayet, O.; Chandler, M. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 1992, 11, 5079–5090. [Google Scholar]
- Kim, J.F.; Charkowski, A.O.; Alfano, J.R.; Collmer, A.; Beer, S.V. Sequences related to transposable elements and bacteriophages flank avirulence genes of Pseudomonas syringae. Mol. Plant Microbe Interact. 1998, 11, 1247–1252. [Google Scholar] [CrossRef]
- Alarcón-Chaidez, F.J.; Peňaloza-Vázquez, A.; Ullrich, M.; Bender, C.L. Characterization of plasmids encoding the phytotoxin coronatine in Pseudomonas syringae. Plasmid 1999, 42, 210–220. [Google Scholar] [CrossRef]
- Lindeberg, M.; Myers, C.R.; Collmer, A.; Schneider, D.J. Roadmap to new virulence determinants in Pseudomonas syringae: Insights from comparative genomics and genome organization. Mol. Plant Microbe Interact. 2008, 21, 685–700. [Google Scholar] [CrossRef]
- Chavan, M.; Rafi, H.; Wertz, J.; Goldstone, C.; Riley, M.A. Phage associated bacteriocins reveal a novel mechanism for bacteriocin diversification in Klebsiella. J. Mol. Evol. 2005, 60, 546–556. [Google Scholar] [CrossRef]
- Hattman, S. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Therapeut. 1999, 84, 367–388. [Google Scholar]
- Charlier, D.; Piette, J.; Glansdorff, N. IS3 can function as a mobile promoter in E.coli. Nucleic Acids Res. 1982, 10, 5935–5948. [Google Scholar] [CrossRef]
- Berg, C.M.; Berg, D.E. Transposable Elements as Tools for Molecular Analysis in Bacteria. In Mobile Genetic Elements: Frontiers in Molecular Biology; Sherratt, D., Ed.; Oxford University Press: Oxford, UK, 1994; pp. 38–68. [Google Scholar]
- Braid, M.D.; Silhavy, J.L.; Kitts, C.L.; Cano, R.J; Howe, M.M. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J. Bacteriol. 2004, 186, 6560–6574. [Google Scholar]
- Morgan, G.J.; Hatfull, G.F.; Casjens, S.; Hendrix, R.W. Bacteriophage Mu genome sequence: Analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 2002, 317, 337–359. [Google Scholar] [CrossRef]
- Witkowski, R.T.; Hattman, S.; Newman, L.; Clark, K.; Tierney, D.L.; Penner-Hahn, J.; McLendon, G. The zinc coordination site of the bacteriophage Mu translational activator protein, Com. J. Mol. Biol. 1995, 247, 753–764. [Google Scholar]
- Kasana, R.C.; Salwan, R.; Dhar, H.; Dutt, S.; Gulati, A. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s Iodine. Curr. Biol. 2008, 57, 503–507. [Google Scholar]
- Baye, E.A.; Chanzyt, H.; Lamed, R.; Shoham, Y. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 1998, 8, 548–557. [Google Scholar]
- Hanekamp, T.; Kobayashi, D.; Hayes, S.; Stayton, M.M. Avirulence gene D of Pseudomonas syringae pv. tomato may have undergone horizontal gene transfer. FEBS Lett. 1997, 415, 40–44. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Palmer, D.A.; Bender, C.L. Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl. Environ. Microbiol. 1993, 59, 1619–1623. [Google Scholar]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar]
- Gerhardt, P.; Murray, R.G.E.; Wood, W.A.; Krieg, N.R. Methods for General and Molecular Bacteriology; American Society for Microbiology: Washington, DC, USA, 1994. [Google Scholar]
- Figurski, D.H.; Helinski, D.R. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar]
- Schenk, A.; Berger, M.; Keith, L.M.; Bender, C.L.; Muskhelishvili, G.; Ullrich, M.S. The algT gene of Pseudomonas syringae pv. glycinea and new insights into the transcriptional organization of the algT-muc gene cluster. J. Bacteriol. 2006, 188, 8013–8021. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, W.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar]
- Green, S.; Studholme, D.J.; Laue, B.E.; Dorati, F.; Lovell, H.; Arnold, D.; Cottrell, J.E.; Bridgett, S.; Blaxter, M.; Huitema, E.; et al. Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum. PLoS One 2011, 5. [Google Scholar]
- Baltrus, D.A.; Nishimura, M.T.; Romanchuk, A.; Chang, J.H.; Mukhtar, M.S.; Cherkis, K.; Roach, J.; Grant, S.R.; Jones, C.D.; Dangl, J.L. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae Isolates. PLoS Pathog. 2011, 7. [Google Scholar]
- Seo, J.W.; Song, K.B.; Jang, K.H.; Kim, C.H.; Jung, B.H.; Rhee, S.K. Molecular cloning of a gene encoding the thermoactive levansucrase from Rahnella aquatilis and its growth phase-dependent expression in Escherichia coli. J. Biotechnol. 2000, 81, 63–72. [Google Scholar]
- Du, Z.; Jakovljevic, V.; Salm, H.; Geider, K. Creation and genetic restoration of Erwinia amylovora strains with low levan synthesis. Physiol. Mol. Plant Pathol. 2004, 65, 115–122. [Google Scholar] [CrossRef]
- Kube, M.; Migdoll, A.M.; Muller, I.; Kuhl, H.; Beck, A.; Reinhardt, R.; Geider, K. The genome of Erwinia tasmaniensis strain Et1/99, a non-pathogenic bacterium in the genus Erwinia. Environ. Microbiol. 2008, 10, 2211–2222. [Google Scholar] [CrossRef]
- Homepage of ExPASy. Available online: http://expasy.org/ (accessed on 18 January 2012).
Appendix
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Srivastava, A.; Al-Karablieh, N.; Khandekar, S.; Sharmin, A.; Weingart, H.; Ullrich, M.S. Genomic Distribution and Divergence of Levansucrase-Coding Genes in Pseudomonas syringae. Genes 2012, 3, 115-137. https://doi.org/10.3390/genes3010115
Srivastava A, Al-Karablieh N, Khandekar S, Sharmin A, Weingart H, Ullrich MS. Genomic Distribution and Divergence of Levansucrase-Coding Genes in Pseudomonas syringae. Genes. 2012; 3(1):115-137. https://doi.org/10.3390/genes3010115
Chicago/Turabian StyleSrivastava, Abhishek, Nehaya Al-Karablieh, Shaunak Khandekar, Arifa Sharmin, Helge Weingart, and Matthias S. Ullrich. 2012. "Genomic Distribution and Divergence of Levansucrase-Coding Genes in Pseudomonas syringae" Genes 3, no. 1: 115-137. https://doi.org/10.3390/genes3010115




