Conserved Role of Heterotrimeric G Proteins in Plant Defense and Cell Death Progression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Condition
2.2. Transient Overexpression Vectors and Infiltration
2.3. RT-qPCR
2.4. Trypan Blue Staining
2.5. DAB Staining
2.6. flg22-Induced ROS
2.7. Electrolyte Leakage Assay
2.8. Protein Content Assay
2.9. Statistical Analyses and Software
3. Results
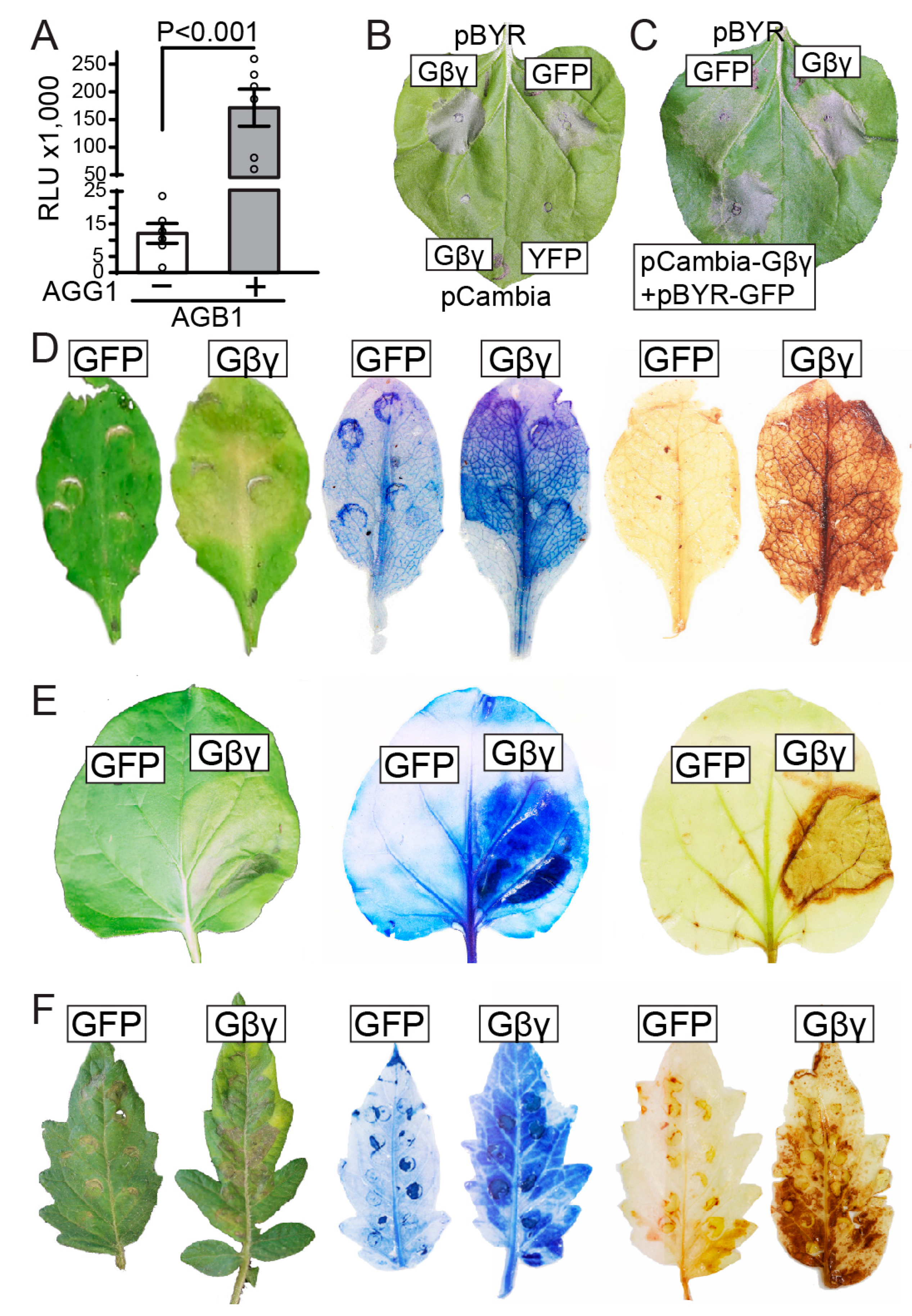
3.1. The Simultaneous Co-Expression of Gβ and Gγ Subunits Significantly Contributed to Cell Death Progression
3.2. Co-Expression of Gβγ Subunits Enhances flg22-Induced ROS Production and Increases Ion Leakage in Arabidopsis and N. benthamiana
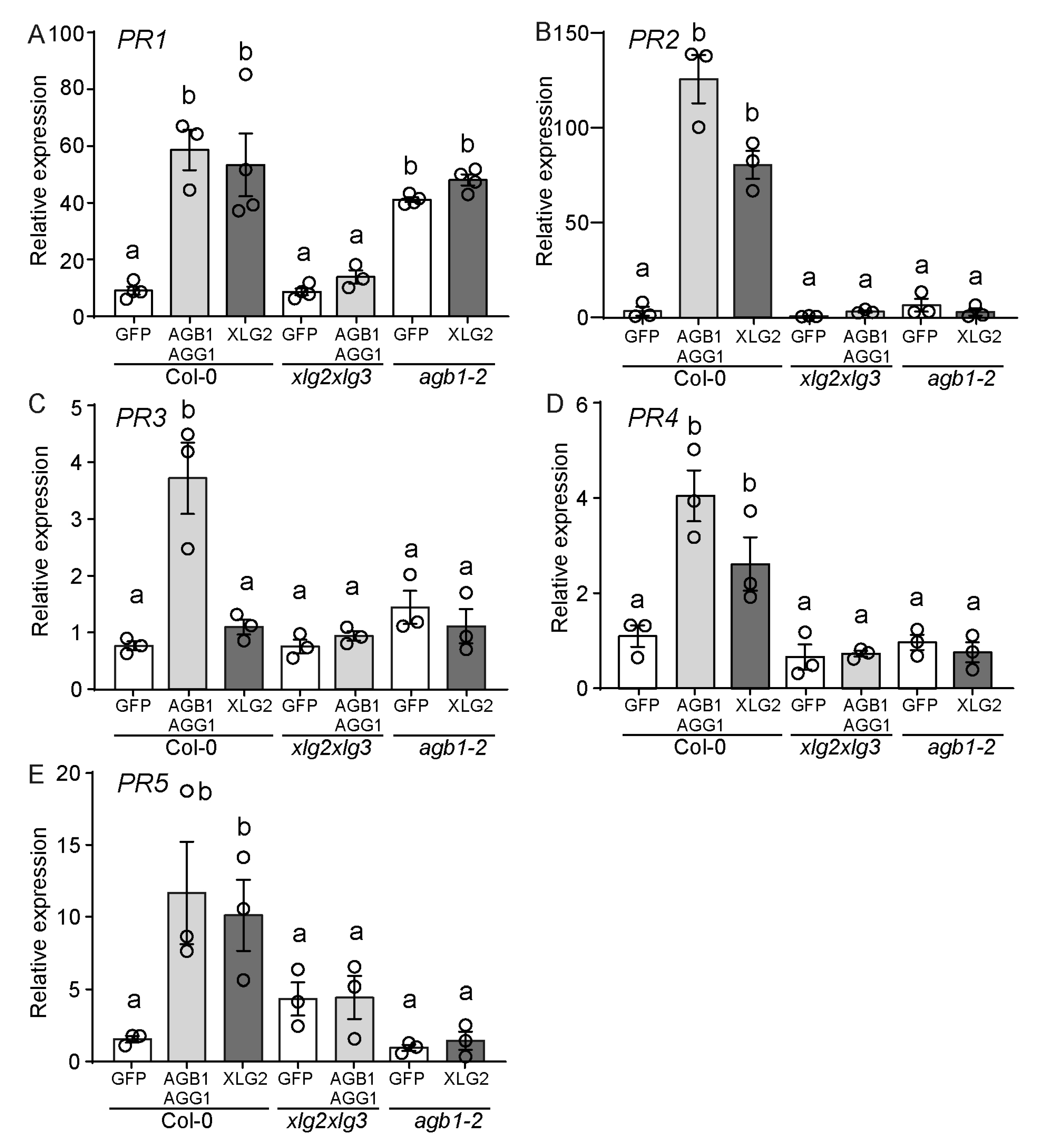
3.3. Gβγ and XLG2 Play Interdependent Roles in the Cell Death Progression
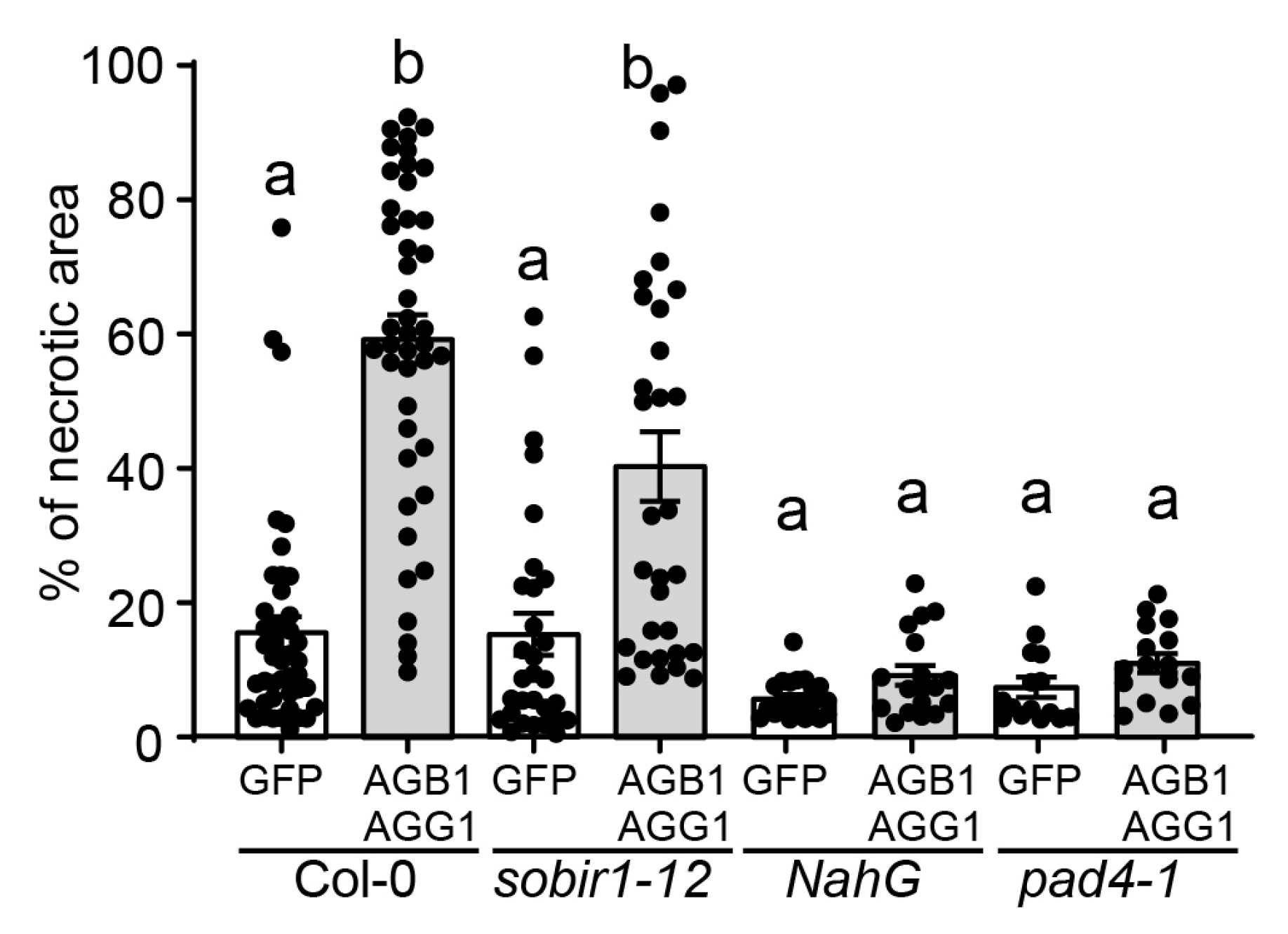
3.4. G Proteins Mediate Cell Death Progression in a Salicylic Acid-Dependent Manner
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Programmed cell death | PCD |
| Heterotrimeric G proteins | G proteins |
| Reactive oxygen species | ROS |
| Receptor-like kinases | RLKs |
| Nucleotide-binding leucine-rich repeat | NLR |
| Arabidopsis G beta 1 | AGB1 |
| Arabidopsis G gamma 1 | AGG1 |
| Extra-large G protein 2 | XLG2 |
| Solanum lycopersicum G beta 1 | SlGB1 |
| Solanum lycopersicum G gamma type A 1 | SlGGA1 |
| BAK1-interacting RLK 1 | BIR1 |
| Suppressor of BIR1 | SOBIR1 |
| Flagellin-Sensitive 2 | FLS2 |
| Enhanced disease susceptibility 1 | EDS1 |
| Phytoalexin deficient 4 | PAD4 |
| Pathogenesis related | PR |
| Salicylic acid | SA |
| Flagellin peptide 22 | flg22 |
| 3′3-Diaminobenzidine | DAB |
| Effector-triggered immunity | ETI |
| High BiT, a part of split luciferase NanoBiT technology developed by Promega Corporation | HiBiT |
| bean yellow dwarf viral | BeYDV |
| Green fluorescent protein | GFP |
| Compact letter display | CLD |
References
- Huysmans, M.; Lema, A.S.; Coll, N.S.; Nowack, M.K. Dying two deaths—Programmed cell death regulation in development and disease. Curr. Opin. Plant Biol. 2017, 35, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.J.; Xiong, J.; Xu, L.T.; Chen, X.W.; Li, W.T. Recent advances in plant immunity with cell death: A review. J. Integr. Agric. 2022, 21, 610–620. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Jones, J.D.G.; Ding, P. Plant immune networks. Trends Plant Sci. 2022, 27, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.T. Programmed Cell Death in Plant-Pathogen Interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 525–545. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Czymmek, K.; Talloczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef]
- Igarashi, D.; Bethke, G.; Xu, Y.; Tsuda, K.; Glazebrook, J.; Katagiri, F. Pattern-triggered immunity suppresses programmed cell death triggered by fumonisin b1. PLoS ONE 2013, 8, e60769. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wen, R.; Wang, J.; Xiang, D.; Wang, Q.; Zang, Y.; Wang, Z.; Huang, S.; Li, X.; Datla, R.; et al. Arabidopsis UBC13 differentially regulates two programmed cell death pathways in responses to pathogen and low-temperature stress. New Phytol. 2018, 221, 919–934. [Google Scholar] [CrossRef] [PubMed]
- Gilman, A.G. G proteins: Transducers of receptor-generated signals. Annu. Rev. Biochem. 1987, 56, 615–649. [Google Scholar] [CrossRef]
- Maruta, N.; Trusov, Y.; Chakravorty, D.; Urano, D.; Assmann, S.M.; Botella, J.R. Nucleotide exchange-dependent and nucleotide exchange-independent functions of plant heterotrimeric GTP-binding proteins. Sci. Signal. 2019, 12, eaav9526. [Google Scholar] [CrossRef] [PubMed]
- Maruta, N.; Trusov, Y.; Urano, D.; Chakravorty, D.; Assmann, S.M.; Jones, A.M.; Botella, J.R. GTP binding by Arabidopsis extra-large G protein 2 is not essential for its functions. Plant Physiol. 2021, 186, 1240–1253. [Google Scholar] [CrossRef] [PubMed]
- Lou, F.; Abramyan, T.M.; Jia, H.; Tropsha, A.; Jones, A.M. An atypical heterotrimeric Gα protein has substantially reduced nucleotide binding but retains nucleotide-independent interactions with its cognate RGS protein and Gβγ dimer. J. Biomol. Struct. Dyn. 2020, 38, 5204–5218. [Google Scholar] [CrossRef] [PubMed]
- Adjobo-Hermans, M.J.; Goedhart, J.; Gadella, T.W. Plant G protein heterotrimers require dual lipidation motifs of Gα and Gγ and do not dissociate upon activation. J. Cell Sci. 2006, 119, 5087–5097. [Google Scholar] [CrossRef] [PubMed]
- Milligan, G.; Kostenis, E. Heterotrimeric G-proteins: A short history. Br. J. Pharmacol. 2006, 147, S46–S55. [Google Scholar] [CrossRef] [PubMed]
- Kankanamge, D.; Tennakoon, M.; Karunarathne, A.; Gautam, N. G protein gamma subunit, a hidden master regulator of GPCR signaling. J. Biol. Chem. 2022, 298, 102618. [Google Scholar] [CrossRef]
- McCudden, C.R.; Willard, F.S.; Kimple, R.J.; Johnston, C.A.; Hains, M.D.; Jones, M.B.; Siderovski, D.P. G alpha selectivity and inhibitor function of the multiple GoLoco motif protein GPSM2/LGN. Biochim. Biophys. Acta 2005, 1745, 254–264. [Google Scholar] [CrossRef]
- Smrcka, A.V. G protein βγ subunits: Central mediators of G protein-coupled receptor signaling. Cell. Mol. Life Sci. 2008, 65, 2191–2214. [Google Scholar] [CrossRef] [PubMed]
- Trusov, Y.; Rookes, J.E.; Tilbrook, K.; Chakravorty, D.; Mason, M.G.; Anderson, D.; Chen, J.G.; Jones, A.M.; Botella, J.R. Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell 2007, 19, 1235–1250. [Google Scholar] [CrossRef]
- Llorente, F.; Alonso-Blanco, C.; Sanchez-Rodriguez, C.; Jorda, L.; Molina, A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. Cell Mol. Biol. 2005, 43, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Ninh, T.T.; Gao, W.; Trusov, Y.; Zhao, J.R.; Long, L.; Song, C.P.; Botella, J.R. Tomato and cotton G protein beta subunit mutants display constitutive autoimmune responses. Plant Direct 2021, 5, e359. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Chen, J.G.; Jones, A.M.; Assmann, S.M. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 2006, 141, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Trusov, Y.; Rookes, J.E.; Chakravorty, D.; Armour, D.; Schenk, P.M.; Botella, J.R. Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol. 2006, 140, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Trusov, Y.; Sewelam, N.; Rookes, J.E.; Kunkel, M.; Nowak, E.; Schenk, P.M.; Botella, J.R. Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant J. Cell Mol. Biol. 2009, 58, 69–81. [Google Scholar] [CrossRef]
- Ullah, H.; Chen, J.G.; Temple, B.; Boyes, D.C.; Alonso, J.M.; Davis, K.R.; Ecker, J.R.; Jones, A.M. The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 2003, 15, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Utsunomiya, Y.; Samejima, C.; Takayanagi, Y.; Izawa, Y.; Yoshida, T.; Sawada, Y.; Fujisawa, Y.; Kato, H.; Iwasaki, Y. Suppression of the rice heterotrimeric G protein beta-subunit gene, RGB1, causes dwarfism and browning of internodes and lamina joint regions. Plant J. Cell Mol. Biol. 2011, 67, 907–916. [Google Scholar] [CrossRef]
- Swain, D.M.; Sahoo, R.K.; Chandan, R.K.; Ghosh, S.; Kumar, R.; Jha, G.; Tuteja, N. Concurrent overexpression of rice G-protein beta and gamma subunits provide enhanced tolerance to sheath blight disease and abiotic stress in rice. Planta 2019, 250, 1505–1520. [Google Scholar] [CrossRef] [PubMed]
- Swain, D.M.; Sahoo, R.K.; Srivastava, V.K.; Tripathy, B.C.; Tuteja, R.; Tuteja, N. Function of heterotrimeric G-protein gamma subunit RGG1 in providing salinity stress tolerance in rice by elevating detoxification of ROS. Planta 2017, 245, 367–383. [Google Scholar] [CrossRef]
- Ding, L.; Pandey, S.; Assmann, S.M. Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J. 2008, 53, 248–263. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Assmann, S.M. Arabidopsis thaliana ’extra-large GTP-binding protein’ (AtXLG1): A new class of G-protein. Plant Mol. Biol. 1999, 40, 55–64. [Google Scholar] [CrossRef]
- Urano, D.; Jones, A.M. Heterotrimeric G protein-coupled signaling in plants. Annu. Rev. Plant Biol. 2014, 65, 365–384. [Google Scholar] [CrossRef]
- Urano, D.; Miura, K.; Wu, Q.; Iwasaki, Y.; Jackson, D.; Jones, A.M. Plant Morphology of Heterotrimeric G Protein Mutants. Plant Cell. Physiol. 2016, 57, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Weiss, C.A.; Garnaat, C.W.; Mukai, K.; Hu, Y.; Ma, H. Isolation of cDNAs encoding guanine nucleotide-binding protein β-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc. Natl. Acad. Sci. USA 1994, 91, 9554–9558. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yanofsky, M.F.; Meyerowitz, E.M. Molecular cloning and characterization of GPA1, a G protein α subunit gene from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1990, 87, 3821–3825. [Google Scholar] [CrossRef]
- Liang, X.; Ding, P.; Lian, K.; Wang, J.; Ma, M.; Li, L.; Li, L.; Li, M.; Zhang, X.; Chen, S.; et al. Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 2016, 5, e13568. [Google Scholar] [CrossRef]
- Liang, X.; Ma, M.; Zhou, Z.; Wang, J.; Yang, X.; Rao, S.; Bi, G.; Li, L.; Zhang, X.; Chai, J.; et al. Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res. 2018, 28, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Maruta, N.; Trusov, Y.; Brenyah, E.; Parekh, U.; Botella, J.R. Membrane-Localized Extra-Large G-Proteins and Gβγ of the Heterotrimeric G Proteins Form Functional Complexes Engaged in Plant Immunity in Arabidopsis. Plant Physiol. 2015, 167, 1004–1016. [Google Scholar] [CrossRef]
- Torres, M.A.; Morales, J.; Sanchez-Rodriguez, C.; Molina, A.; Dangl, J. Functional interplay between Arabidopsis NADPH oxidases and heterotrimeric G protein. Mol. Plant-Microbe Interact. MPMI 2013, 26, 686–694. [Google Scholar] [CrossRef]
- Liang, Y.; Gao, Y.; Jones, A.M. Extra large G-protein interactome reveals multiple stress response function and partner-dependent XLG subcellular localization. Front. Plant Sci. 2017, 8, 1015. [Google Scholar] [CrossRef]
- Petutschnig, E.; Anders, J.; Stolze, M.; Meusel, C.; Hacke, R.; Much, L.; Schwier, M.; Gippert, A.L.; Kroll, S.; Fasshauer, P.; et al. EXTRA LARGE G-PROTEIN2 mediates cell death and hyperimmunity in the chitin elicitor receptor kinase 1-4 mutant. Plant Physiol. 2022, 189, 2413–2431. [Google Scholar] [CrossRef]
- Ishikawa, A. The Arabidopsis G-protein β-subunit is required for defense response against Agrobacterium tumefaciens. Biosci. Biotechnol. Biochem. 2009, 73, 47–52. [Google Scholar] [CrossRef]
- Liu, J.; Ding, P.; Sun, T.; Nitta, Y.; Dong, O.; Huang, X.; Yang, W.; Li, X.; Botella, J.R.; Zhang, Y. Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol. 2013, 161, 2146–2158. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Wang, P.; Zhong, H.; Liu, W.; Zhang, S.; Xiong, L.; Wu, Y.; Xia, Y. Arabidopsis EXTRA-LARGE G PROTEIN 1 (XLG1) functions together with XLG2 and XLG3 in PAMP-triggered MAPK activation and immunity. J. Integr. Plant Biol. 2023, 65, 825–837. [Google Scholar] [CrossRef]
- Gao, Y.; Gu, H.; Leburu, M.; Li, X.; Wang, Y.; Sheng, J.; Fang, H.; Gu, M.; Liang, G. The heterotrimeric G protein beta subunit RGB1 is required for seedling formation in rice. Rice 2019, 12, 53. [Google Scholar] [CrossRef]
- Urano, D.; Leong, R.; Wu, T.Y.; Jones, A.M. Quantitative morphological phenomics of rice G protein mutants portend autoimmunity. Dev. Biol. 2020, 457, 83–90. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, F.; Liu, L.; Char, S.N.; Ding, Y.; Je, B.I.; Schmelz, E.; Yang, B.; Jackson, D. The maize heterotrimeric G protein β subunit controls shoot meristem development and immune responses. Proc. Natl. Acad. Sci. USA 2020, 117, 1799–1805. [Google Scholar] [CrossRef]
- Ullah, H.; Chen, J.G.; Wang, S.; Jones, A.M. Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 2002, 129, 897–907. [Google Scholar] [CrossRef]
- Zhu, H.; Li, G.J.; Ding, L.; Cui, X.; Berg, H.; Assmann, S.M.; Xia, Y. Arabidopsis extra large G-protein 2 (XLG2) interacts with the Gβ subunit of heterotrimeric G protein and functions in disease resistance. Mol. Plant 2009, 2, 513–525. [Google Scholar] [CrossRef]
- Pandey, S.; Monshausen, G.B.; Ding, L.; Assmann, S.M. Regulation of root-wave response by extra large and conventional G proteins in Arabidopsis thaliana. Plant J. 2008, 55, 311–322. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Manners, J.M.; Kazan, K. Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. Cell Mol. Biol. 2009, 58, 927–939. [Google Scholar] [CrossRef]
- Glazebrook, J.; Rogers, E.E.; Ausubel, F.M. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 1996, 143, 973–982. [Google Scholar] [CrossRef]
- Diamos, A.G.; Rosenthal, S.H.; Mason, H.S. 5′ and 3′ Untranslated Regions Strongly Enhance Performance of Geminiviral Replicons in Nicotiana benthamiana Leaves. Front. Plant Sci. 2016, 7, 200. [Google Scholar] [CrossRef]
- Hatsugai, N.; Katagiri, F. Quantification of Plant Cell Death by Electrolyte Leakage Assay. Bio-Protoc. 2018, 8, e2758. [Google Scholar] [CrossRef] [PubMed]
- Dixon, A.S.; Schwinn, M.K.; Hall, M.P.; Zimmerman, K.; Otto, P.; Lubben, T.H.; Butler, B.L.; Binkowski, B.F.; Machleidt, T.; Kirkland, T.A.; et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408. [Google Scholar] [CrossRef]
- Wolfenstetter, S.; Chakravorty, D.; Kula, R.; Urano, D.; Trusov, Y.; Sheahan, M.B.; McCurdy, D.W.; Assmann, S.M.; Jones, A.M.; Botella, J.R. Evidence for an unusual transmembrane configuration of AGG3, a class C Gγ subunit of Arabidopsis. Plant J. Cell Mol. Biol. 2015, 81, 388–398. [Google Scholar] [CrossRef]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-induced electrolyte leakage: The role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef]
- Leng, J.; Tu, W.; Hou, Y.; Cui, H. Temperature-Inducible Transgenic EDS1 and PAD4 in Arabidopsis Confer an Enhanced Disease Resistance at Elevated Temperature. Plants 2021, 10, 1258. [Google Scholar] [CrossRef] [PubMed]
- Maruta, N.; Burdett, H.; Lim, B.Y.J.; Hu, X.; Desa, S.; Manik, M.K.; Kobe, B. Structural basis of NLR activation and innate immune signalling in plants. Immunogenetics 2022, 74, 5–26. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.H.; Wang, X.; Wang, D.M.; Xu, F.; Ding, X.J.; Zhang, Z.B.; Bi, D.L.; Cheng, Y.T.; Chen, S.; Li, X.; et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 2009, 6, 34–44. [Google Scholar] [CrossRef]
- Takahashi, T.; Murano, T.; Ishikawa, A. SOBIR1 and AGB1 independently contribute to nonhost resistance to Pyricularia oryzae (syn. Magnaporthe oryzae) in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2018, 82, 1922–1930. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Li, L.; Weigel, D. One Hundred Years of Hybrid Necrosis: Hybrid Autoimmunity as a Window into the Mechanisms and Evolution of Plant-Pathogen Interactions. Annu. Rev. Phytopathol. 2021, 59, 213–237. [Google Scholar] [CrossRef]
- Chen, J.G.; Willard, F.S.; Huang, J.; Liang, J.; Chasse, S.A.; Jones, A.M.; Siderovski, D.P. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 2003, 301, 1728–1731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hu, G.; Cheng, Y.; Huang, J. Heterotrimeric G protein α and β subunits antagonistically modulate stomatal density in Arabidopsis thaliana. Dev. Biol. 2008, 324, 68–75. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karimian, P.; Trusov, Y.; Botella, J.R. Conserved Role of Heterotrimeric G Proteins in Plant Defense and Cell Death Progression. Genes 2024, 15, 115. https://doi.org/10.3390/genes15010115
Karimian P, Trusov Y, Botella JR. Conserved Role of Heterotrimeric G Proteins in Plant Defense and Cell Death Progression. Genes. 2024; 15(1):115. https://doi.org/10.3390/genes15010115
Chicago/Turabian StyleKarimian, Parastoo, Yuri Trusov, and Jose Ramon Botella. 2024. "Conserved Role of Heterotrimeric G Proteins in Plant Defense and Cell Death Progression" Genes 15, no. 1: 115. https://doi.org/10.3390/genes15010115




