Nonribosomal Peptide Synthetases in Animals
Abstract
:1. Introduction
2. Materials and Methods
2.1. NRPS Identification
2.2. Phylogenetic Analyses
2.3. NRPSs in Rotifers
2.4. Beta-Lactam Biosynthesis Genes in Plectus Sambesii
3. Results
3.1. The Distribution of NRPS Genes in Animals
3.2. Putative NRPSs Are Abundant in Rotifers
3.3. Beta-Lactam Biosynthesis Genes in P. sambesii
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264. [Google Scholar] [CrossRef] [PubMed]
- Shou, Q.; Feng, L.; Long, Y.; Han, J.; Nunnery, J.K.; Powell, D.H.; A Butcher, R. A hybrid polyketide-nonribosomal peptide in nematodes that promotes larval survival. Nat. Chem. Biol. 2016, 12, 770–772. [Google Scholar] [CrossRef] [PubMed]
- Roelofs, D.; Timmermans, M.J.T.N.; Hensbergen, P.; Van Leeuwen, H.; Koopman, J.; Faddeeva, A.; Suring, W.; de Boer, T.E.; Mariën, J.; Boer, R.; et al. A functional isopenicillin N synthase in an animal genome. Mol. Biol. Evol. 2013, 30, 541–548. [Google Scholar] [CrossRef]
- Chen, R.; Wang, J.; Gradinaru, I.; Vu, H.S.; Geboers, S.; Naidoo, J.; Ready, J.M.; Williams, N.S.; DeBerardinis, R.J.; Ross, E.M.; et al. A male-derived nonribosomal peptide pheromone controls female schistosome development. Cell 2022, 185, 1506–1520. [Google Scholar] [CrossRef] [PubMed]
- O’brien, R.V.; Davis, R.W.; Khosla, C.; Hillenmeyer, M.E. Computational identification and analysis of orphan assembly-line polyketide synthases. J. Antibiot. 2014, 67, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Suring, W.; Meusemann, K.; Blanke, A.; Mariën, J.; Schol, T.; Agamennone, V.; Faddeeva-Vakhrusheva, A.; Berg, M.P.; Brouwer, A.; van Straalen, N.M.; et al. Evolutionary ecology of beta-lactam gene clusters in animals. Mol. Ecol. 2017, 26, 3217–3229. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; Gulick, A.M. Structural biology of nonribosomal peptide synthetases. Methods Mol. Biol. 2016, 1401, 3–29. [Google Scholar] [PubMed]
- Flissi, A.; Ricart, E.; Campart, C.; Chevalier, M.; Dufresne, Y.; Michalik, J.; Jacques, P.; Flahaut, C.; Lisacek, F.; Leclère, V.; et al. Norine: Update of the nonribosomal peptide resource. Nucleic Acids Res. 2020, 48, D465–D469. [Google Scholar] [PubMed]
- Lambalot, R.H.; Gehring, A.M.; Flugel, R.S.; Zuber, P.; LaCelle, M.; Marahiel, M.A.; Reid, R.; Khosla, C.; Walsh, C.T. A new enzyme superfamily—The phosphopantetheinyl transferases. Chem. Biol. 1996, 3, 923–936. [Google Scholar] [CrossRef]
- Mootz, H.D.; Finking, R.; Marahiel, M.A. 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 2001, 276, 37289–37298. [Google Scholar] [CrossRef]
- Suring, W.; Mariën, J.; Broekman, R.; Van Straalen, N.M.; Roelofs, D. Biochemical pathways supporting beta-lactam biosynthesis in the springtail Folsomia candida. Biol. Open 2016, 5, 1784–1789. [Google Scholar] [CrossRef]
- Richardt, A.; Kemme, T.; Wagner, S.; Schwarzer, D.; Marahiel, M.A.; Hovemann, B.T. Ebony, a novel nonribosomal peptide synthetase for β-alanine conjugation with biogenic amines in Drosophila. J. Biol. Chem. 2003, 278, 41160–41166. [Google Scholar] [CrossRef] [PubMed]
- Drozak, J.; Veiga-da-Cunha, M.; Kadziolka, B.; Van Schaftingen, E. Vertebrate Acyl CoA synthetase family member 4 (ACSF 4-U26) is a β-alanine-activating enzyme homologous to bacterial non-ribosomal peptide synthetase. FEBS J. 2014, 281, 1585–1597. [Google Scholar] [CrossRef]
- Izoré, T.; Tailhades, J.; Hansen, M.H.; Kaczmarski, J.A.; Jackson, C.J.; Cryle, M.J. Drosophila melanogaster nonribosomal peptide synthetase Ebony encodes an atypical condensation domain. Proc. Natl. Acad. Sci. USA 2019, 116, 2913–2918. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; True, J.R.; Carroll, S.B. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development 2002, 129, 1849–1858. [Google Scholar] [CrossRef]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; De Los Santos, E.L.; Kim, H.U.; Nave, M.; et al. AntiSMASH 4.0—Improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]
- Hinchliff, C.E.; Smith, S.A.; Allman, J.F.; Burleigh, J.G.; Chaudhary, R.; Coghill, L.M.; Crandall, K.A.; Deng, J.; Drew, B.T.; Gazis, R.; et al. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc. Natl. Acad. Sci. USA 2015, 112, 12764–12769. [Google Scholar] [CrossRef] [PubMed]
- Michonneau, F.; Brown, J.W.; Winter, D.J. rotl: An R package to interact with the Open Tree of Life data. Methods Ecol. Evol. 2016, 7, 1476–1481. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Ahmed, M.; Roberts, N.G.; Adediran, F.; Smythe, A.B.; Kocot, K.M.; Holovachov, O. Phylogenomic Analysis of the Phylum Nematoda: Conflicts and Congruences with Morphology, 18S rRNA, and Mitogenomes. Front. Ecol. Evol. 2022, 9, 769565. [Google Scholar] [CrossRef]
- Simion, P.; Narayan, J.; Houtain, A.; Derzelle, A.; Baudry, L.; Nicolas, E.; Arora, R.; Cariou, M.; Cruaud, C.; Gaudray, F.R.; et al. Chromosome-level genome assembly reveals homologous chromosomes and recombination in asexual rotifer Adineta vaga. Sci. Adv. 2021, 7, eabg4216. [Google Scholar] [CrossRef]
- Rebecchi, L.; Cesari, M.; Altiero, T.; Frigieri, A.; Guidetti, R. Survival and DNA degradation in anhydrobiotic tardigrades. J. Exp. Biol. 2009, 212, 4033–4039. [Google Scholar] [CrossRef]
- Barra, J.A.; Poinsot-Balaguer, N. Modifications ultrastructurales accompagnant l’anhydrobiose chez un Collembole: Folsomides variabilis. Rev. Ecol. Biol. Sol 1977, 14, 189–197. [Google Scholar]
- Gladyshev, E.; Meselson, M. Extreme resistance of bdelloid rotifers to ionizing radiation. Proc. Natl. Acad. Sci. USA 2008, 105, 5139–5144. [Google Scholar] [CrossRef]
- Crisp, A.; Boschetti, C.; Perry, M.; Tunnacliffe, A.; Micklem, G. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 2015, 16, 50. [Google Scholar] [CrossRef]
- Gladyshev, E.A.; Arkhipova, I.R. Genome structure of bdelloid rotifers: Shaped by asexuality or desiccation? J. Hered. 2010, 101 (Suppl. S1), S85–S93. [Google Scholar] [CrossRef] [PubMed]
- Hespeels, B.; Knapen, M.; Hanot-Mambres, D.; Heuskin, A.; Pineux, F.; Lucas, S.; Koszul, R.; van Doninck, K. Gateway to genetic exchange? DNA double-strand breaks in the bdelloid rotifer Adineta vaga submitted to desiccation. J. Evol. Biol. 2014, 27, 1334–1345. [Google Scholar] [CrossRef] [PubMed]
- Eyres, I.; Boschetti, C.; Crisp, A.; Smith, T.P.; Fontaneto, D.; Tunnacliffe, A.; Barraclough, T.G. Horizontal gene transfer in bdelloid rotifers is ancient, ongoing and more frequent in species from desiccating habitats. BMC Biol. 2015, 13, 90. [Google Scholar] [CrossRef]
- Bushley, K.E.; Turgeon, B.G. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol. Biol. 2010, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.; Schneider, P.; Tosch, W.; Scartazzini, R.; Zak, O. The penems, a new class of β-lactam antibiotics. 7. Synthesis and antimicrobial activity of 2-heterocyclylmercaptoalkyl derivatives. J. Antibiot. 1986, 39, 525–534. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Fierro, F.; Casqueiro, J.; Martín, J.F. Gene organization and plasticity of the β-lactam genes in different filamentous fungi. Antonie Van Leeuwenhoek 1999, 75, 81–94. [Google Scholar] [CrossRef]
- García-Estrada, C.; Vaca, I.; Ullán, R.V.; Van Den Berg, M.A.; Bovenberg RAl Martín, J.F. Molecular characterization of a fungal gene paralogue of the penicillin penDE gene of Penicillium chrysogenum. BMC Microbiol. 2009, 9, 104. [Google Scholar] [CrossRef]
- Torres, J.P.; Schmidt, E.W. The biosynthetic diversity of the animal world. J. Biol. Chem. 2019, 294, 17684–17692. [Google Scholar] [CrossRef]


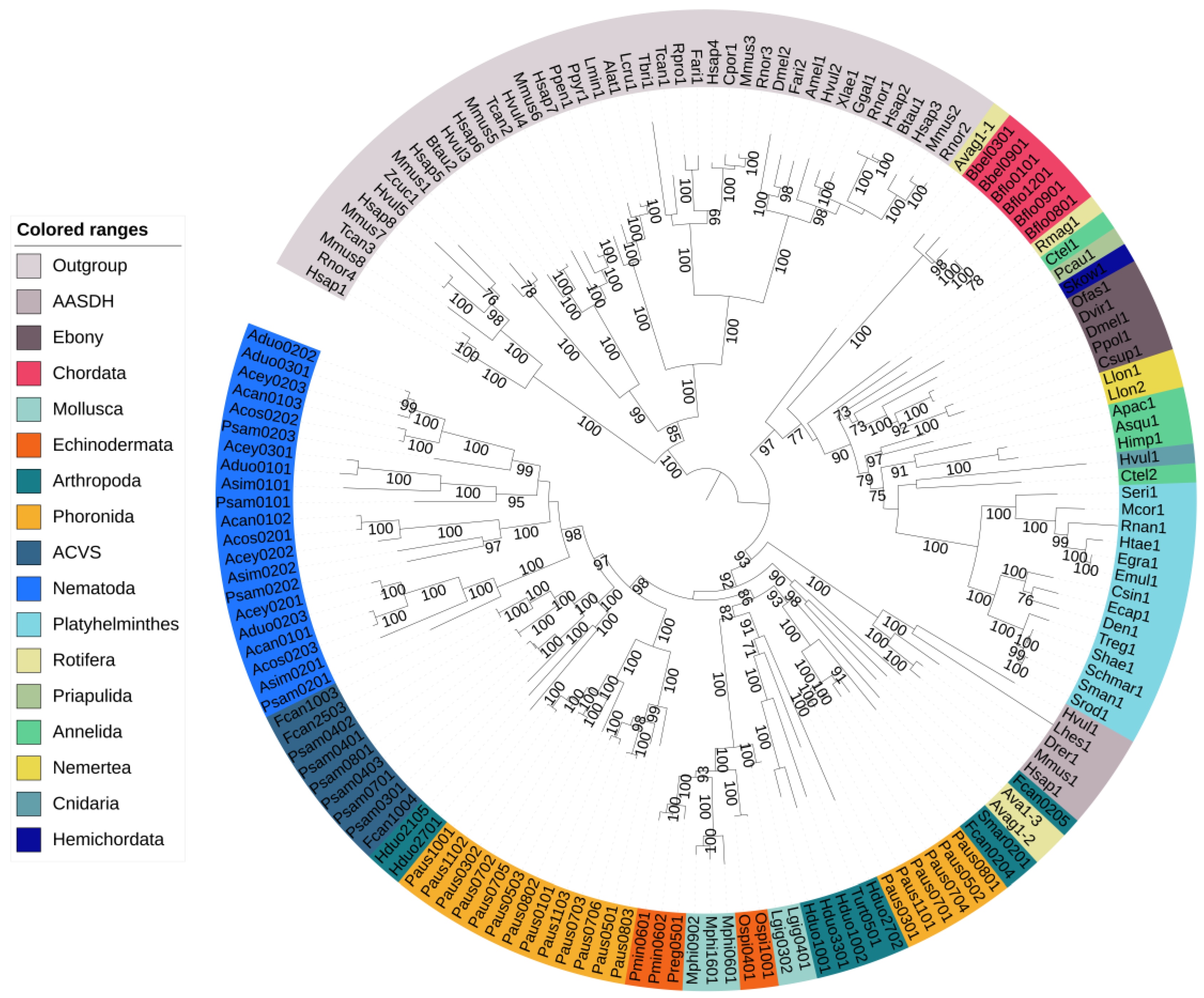
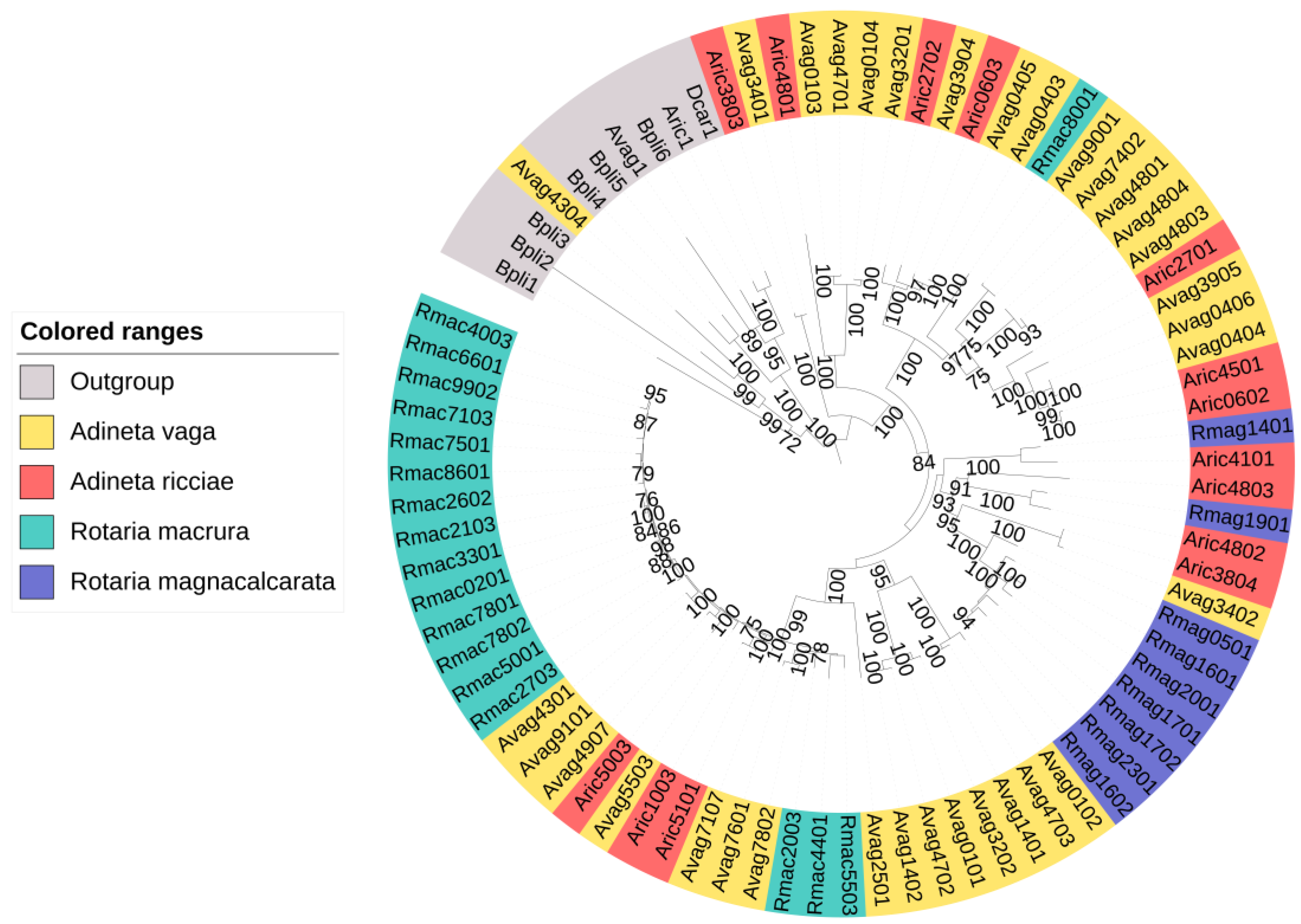
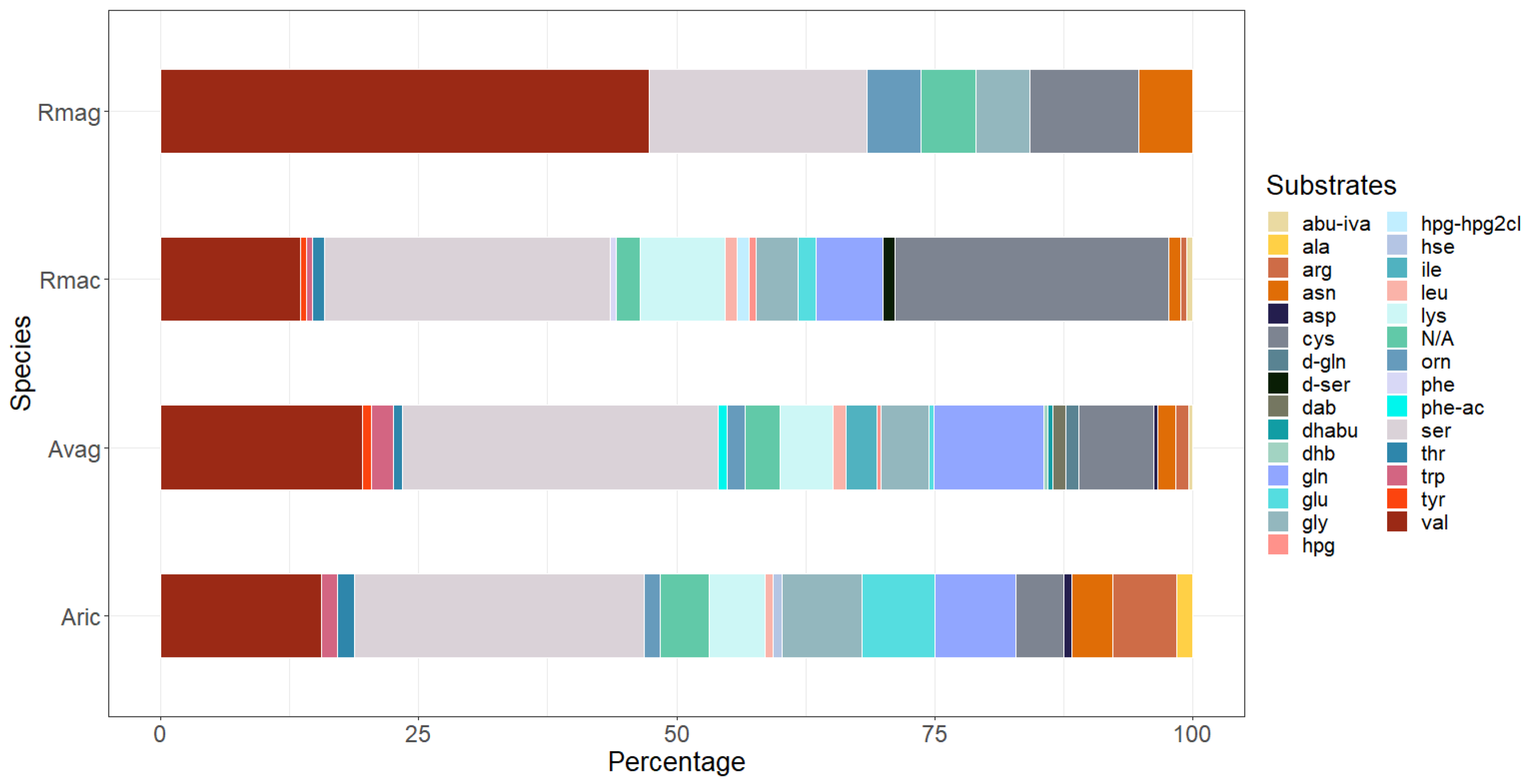

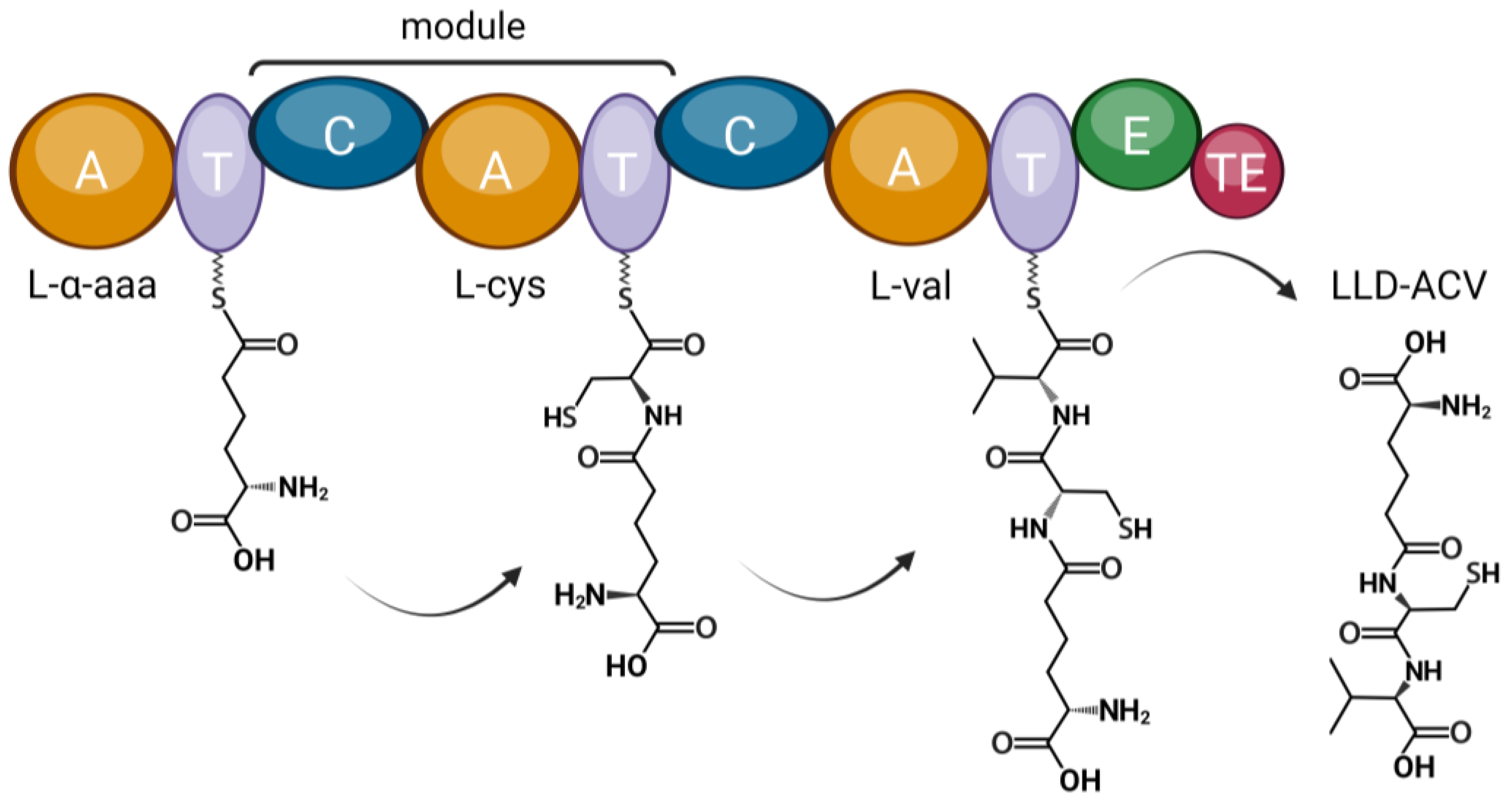
| Species | NRPS | NRPS–PKS |
|---|---|---|
| Adineta ricciae | 46 | 2 |
| Adineta vaga | 69 | 5 |
| Rotaria macrura | 79 | 0 |
| Rotaria magnacalcarata | 12 | 1 |
| Brachionus calyciflorus | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suring, W.; Hoogduin, D.; Le Ngoc, G.; Brouwer, A.; van Straalen, N.M.; Roelofs, D. Nonribosomal Peptide Synthetases in Animals. Genes 2023, 14, 1741. https://doi.org/10.3390/genes14091741
Suring W, Hoogduin D, Le Ngoc G, Brouwer A, van Straalen NM, Roelofs D. Nonribosomal Peptide Synthetases in Animals. Genes. 2023; 14(9):1741. https://doi.org/10.3390/genes14091741
Chicago/Turabian StyleSuring, Wouter, Dylan Hoogduin, Giang Le Ngoc, Abraham Brouwer, Nico M. van Straalen, and Dick Roelofs. 2023. "Nonribosomal Peptide Synthetases in Animals" Genes 14, no. 9: 1741. https://doi.org/10.3390/genes14091741






