Small RNA Profiling of Aster Yellows Phytoplasma-Infected Catharanthus roseus Plants Showing Different Symptoms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phytoplasma Detection and Quantification
2.3. Small RNA HTS
2.4. Bioinformatic Analysis of the Small RNA Sequences
2.5. Small RNA Northern Blot
2.6. Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis of the Predicted Target Genes
3. Results
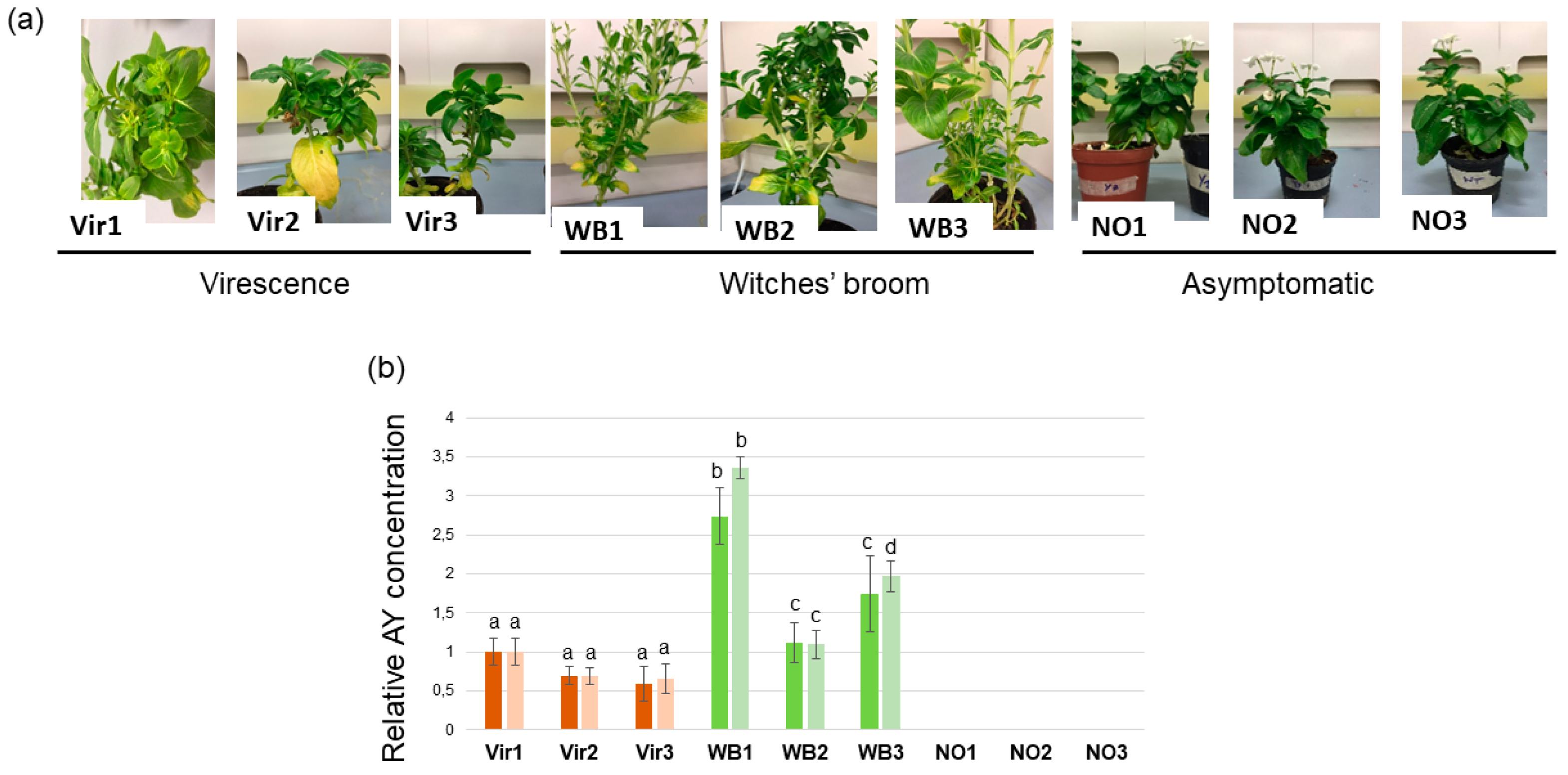
3.1. Phytoplasma Concentration in Periwinkle Plants
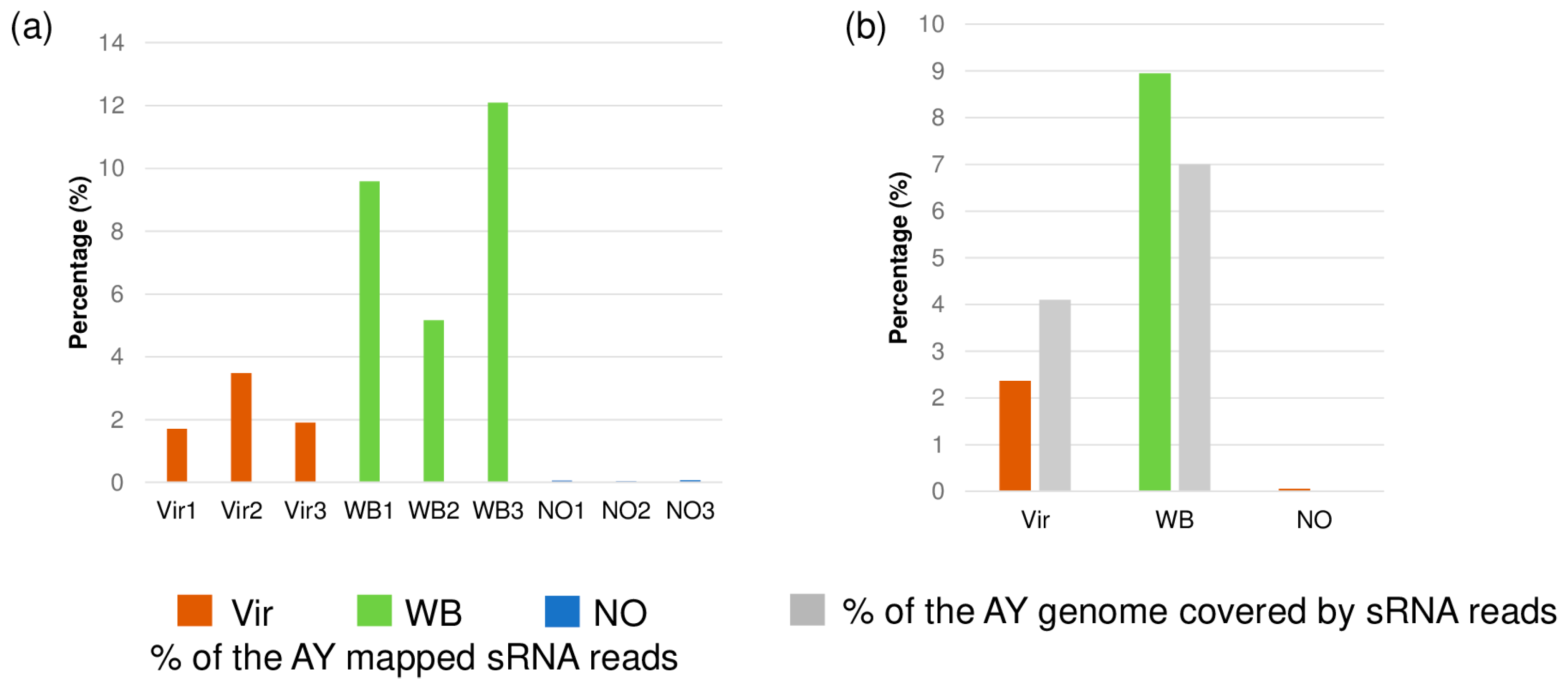
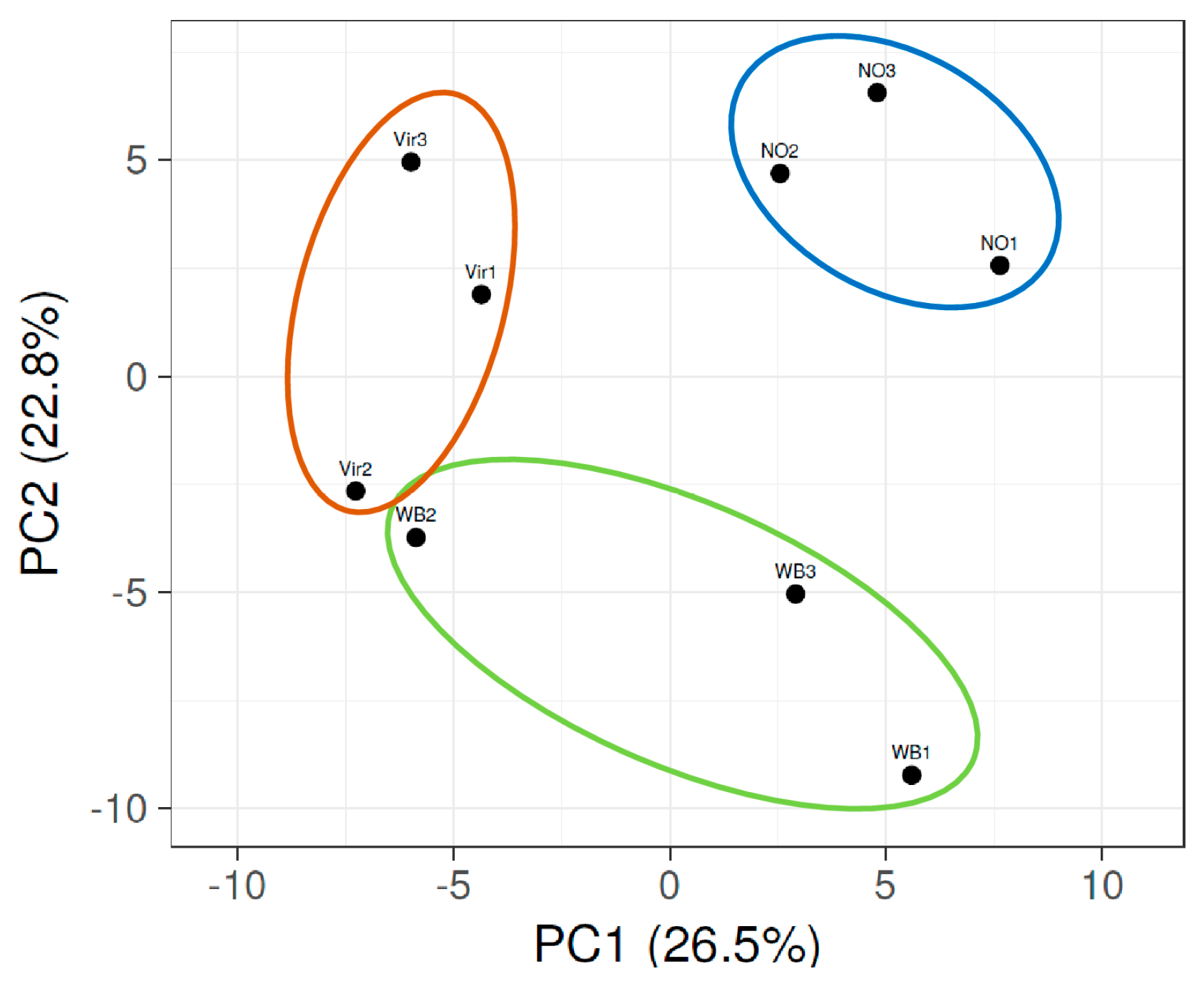
3.2. Small RNA HTS Profiling
3.3. Characteristics of Phytoplasma-Derived Small RNAs
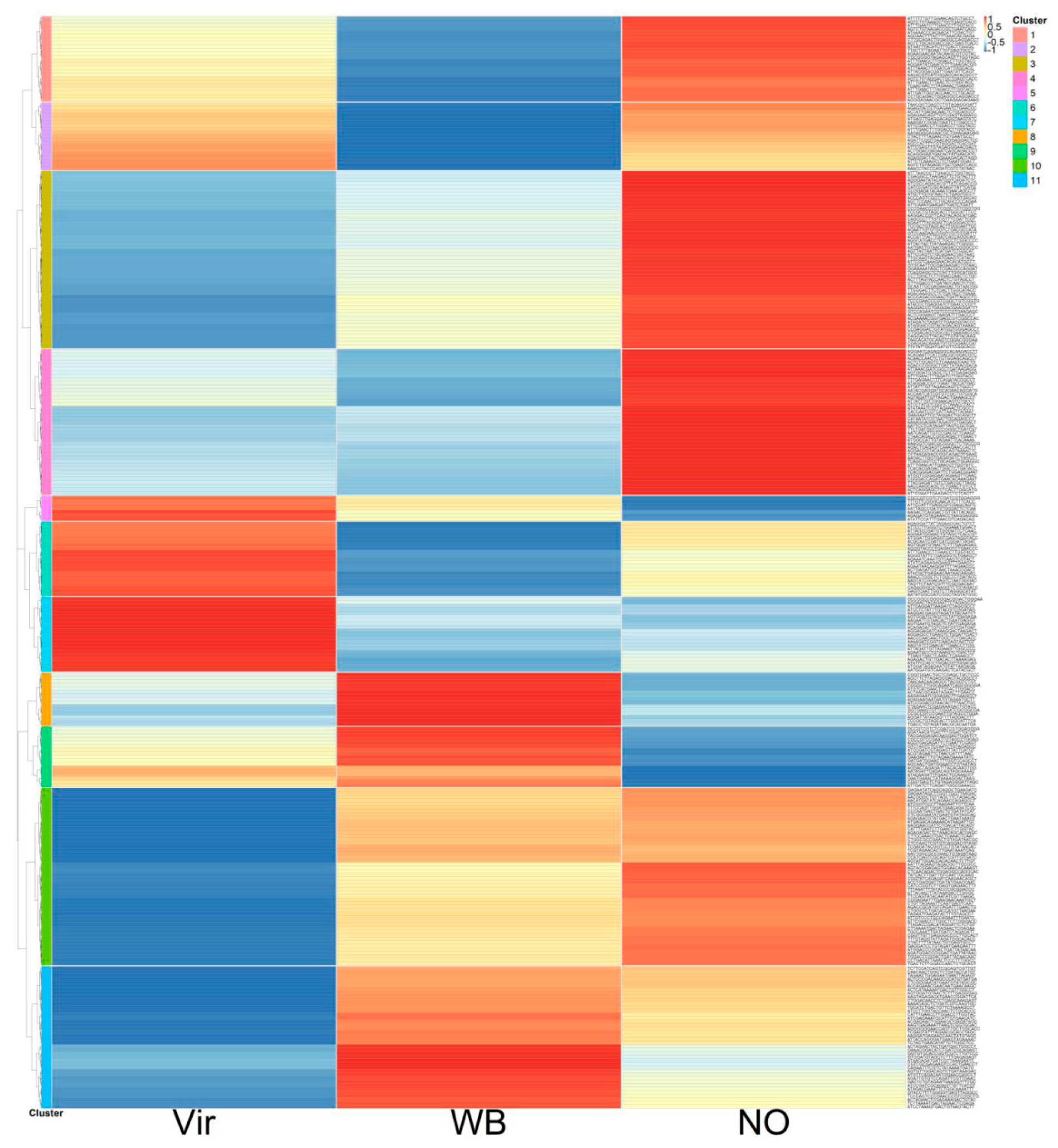
3.4. The miRNA Profile of the Plants, Showing Different Symptoms
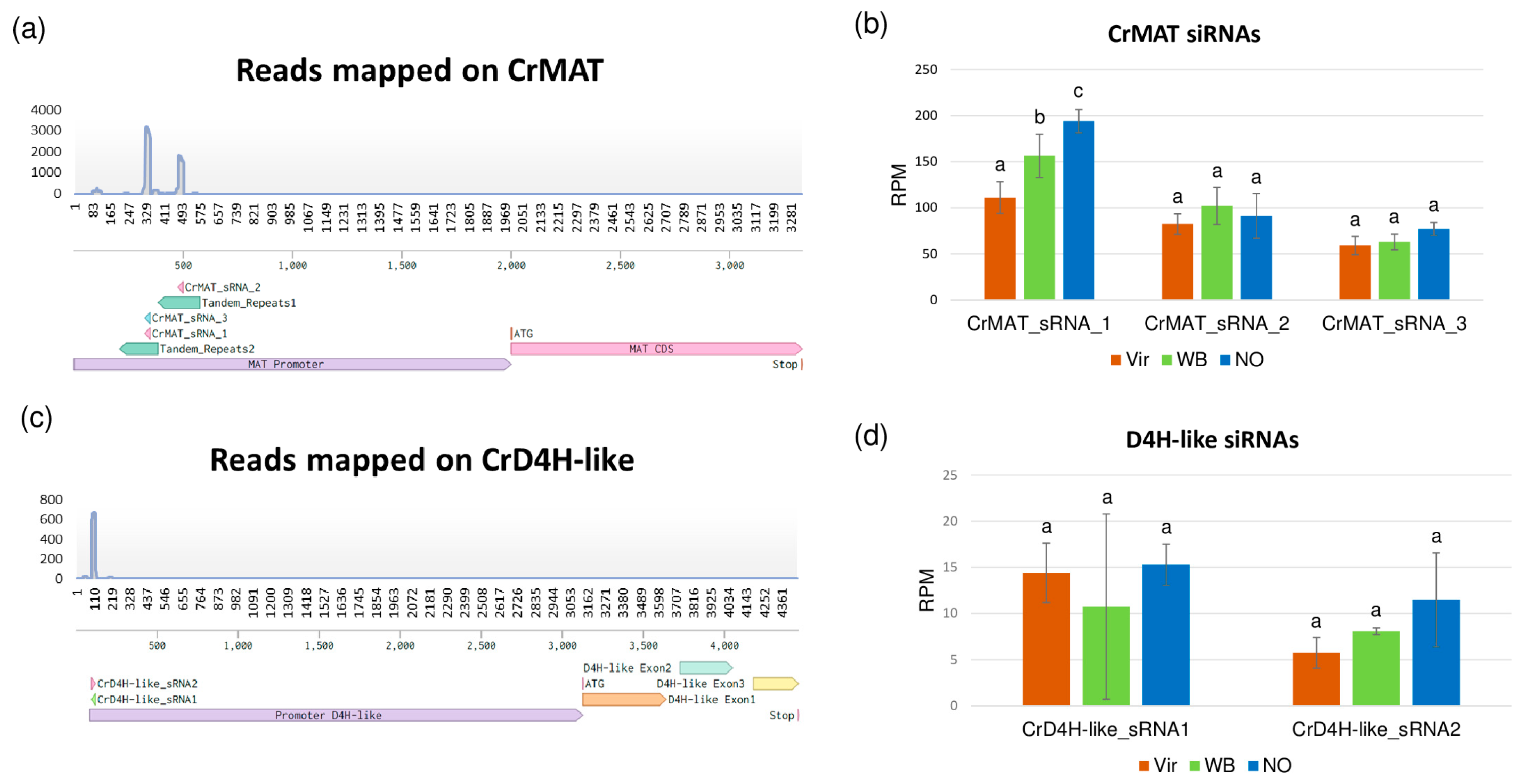
3.5. The siRNA Profiles of the Investigated Plants
3.6. Investigation of the TIA Pathway Key Regulator Expression during Phytoplasma Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- IRPCM. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 2004, 54, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Bertaccini, A. Plants and Phytoplasmas: When Bacteria Modify Plants. Plants 2022, 11, 1425. [Google Scholar] [CrossRef] [PubMed]
- Dermastia, M. Plant Hormones in Phytoplasma Infected Plants. Front. Plant Sci. 2019, 10, 477. [Google Scholar] [CrossRef] [PubMed]
- Bendix, C.; Lewis, J.D. The enemy within: Phloem-limited pathogens. Mol. Plant Pathol. 2018, 19, 238–254. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005, 579, 5923–5931. [Google Scholar] [CrossRef]
- Szittya, G.; Burgyán, J. RNA interference-mediated intrinsic antiviral immunity in plants. Curr. Top Microbiol. Immunol. 2013, 371, 153–181. [Google Scholar]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef]
- Wang, L.; Chen, W.; Ma, H.; Li, J.; Hao, X.; Wu, Y. Identification of RNA silencing suppressor encoded by wheat blue dwarf (WBD) phytoplasma. Plant Biol. 2021, 23, 843–849. [Google Scholar] [CrossRef]
- Kaviani, M.; Goodwin, P.H.; Hunter, D.M. Differences in Gene Expression of Pear Selections Showing Leaf Curling or Leaf Reddening Symptoms Due to Pear Decline Phytoplasma. Plants 2022, 11, 427. [Google Scholar] [CrossRef]
- Tan, Y.; Li, Q.; Zhao, Y.; Wei, H.; Wang, J.; Baker, C.J.; Liu, Q.; Wei, W. Integration of metabolomics and existing omics data reveals new insights into phytoplasma-induced metabolic reprogramming in host plants. PLoS ONE 2021, 16, e0246203. [Google Scholar] [CrossRef]
- Wang, X.; Hu, Q.; Wang, J.; Lou, L.; Xu, X.; Chen, X. Comparative Biochemical and Transcriptomic Analyses Provide New Insights into Phytoplasma Infection Responses in Cucumber. Genes 2022, 13, 1903. [Google Scholar] [CrossRef] [PubMed]
- Dermastia, M.; Škrlj, B.; Strah, R.; Anžič, B.; Tomaž, Š.; Križnik, M.; Schönhuber, C.; Riedle-Bauer, M.; Ramšak, Ž.; Petek, M.; et al. Differential Response of Grapevine to Infection with ‘Candidatus Phytoplasma solani’ in Early and Late Growing Season through Complex Regulation of mRNA and Small RNA Transcriptomes. Int. J. Mol. Sci. 2021, 22, 3531. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Dong, X.; Xu, Y.; Dong, Q.; Wang, Y.; Gai, Y.; Ji, X. Transcriptome and DNA Methylome Reveal Insights Into Phytoplasma Infection Responses in Mulberry (Morus multicaulis Perr.). Front. Plant Sci. 2021, 12, 697702. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhai, X.; Zhao, Z.; Deng, M.; Li, Y.; Fan, G. Genome-wide DNA methylation analysis of paulownia with phytoplasma infection. Gene 2020, 755, 144905. [Google Scholar] [CrossRef]
- Ehya, F.; Monavarfeshani, A.; Mohseni Fard, E.; Karimi Farsad, L.; Khayam Nekouei, M.; Mardi, M.; Salekdeh, G.H. Phytoplasma-Responsive microRNAs Modulate Hormonal, Nutritional, and Stress Signalling Pathways in Mexican Lime Trees. PLoS ONE 2013, 8, e66372. [Google Scholar] [CrossRef]
- Gai, Y.P.; Li, Y.Q.; Guo, F.Y.; Yuan, C.Z.; Mo, Y.Y.; Zhang, H.L.; Wang, H.; Ji, X.L. Analysis of phytoplasma-responsive sRNAs provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 2014, 4, 5378. [Google Scholar] [CrossRef]
- Fan, G.; Cao, X.; Niu, S.; Deng, M.; Zhao, Z.; Dong, Y. Transcriptome, microRNA, and degradome analyses of the gene expression of Paulownia with phytoplamsa. BMC Genom. 2015, 16, 896. [Google Scholar] [CrossRef]
- Fan, G.; Niu, S.; Xu, T.; Deng, M.; Zhao, Z.; Wang, Y.; Cao, L.; Wang, Z. Plant-Pathogen Interaction-Related MicroRNAs and Their Targets Provide Indicators of Phytoplasma Infection in Paulownia tomentosa × Paulownia fortunei. PLoS ONE 2015, 10, e0140590. [Google Scholar] [CrossRef]
- Shao, F.; Zhang, Q.; Liu, H.; Lu, S.; Qiu, D. Genome-Wide Identification and Analysis of MicroRNAs Involved in Witches’-Broom Phytoplasma Response in Ziziphus jujuba. PLoS ONE 2016, 11, e0166099. [Google Scholar] [CrossRef]
- Snyman, M.C.; Solofoharivelo, M.C.; Souza-Richards, R.; Stephan, D.; Murray, S.; Burger, J.T. The use of high-throughput small RNA sequencing reveals differentially expressed microRNAs in response to aster yellows phytoplasma-infection in Vitis vinifera cv. ‘Chardonnay’. PLoS ONE 2017, 12, e0182629. [Google Scholar] [CrossRef]
- Chitarra, W.; Pagliarani, C.; Abbà, S.; Boccacci, P.; Birello, G.; Rossi, M.; Palmano, S.; Marzachì, C.; Perrone, I.; Gambino, G. miRVIT: A Novel miRNA Database and Its Application to Uncover Vitis Responses to Flavescence dorée Infection. Front. Plant Sci. 2018, 9, 1034. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xing, S.; Pan, Q.; Yuan, F.; Zhao, J.; Tian, Y.; Chen, Y.; Wang, G.; Tang, K. Development of efficient Catharanthus roseus regeneration and transformation system using agrobacterium tumefaciens and hypocotyls as explants. BMC Biotechnol. 2012, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Leopold, A.L.; Sander, G.W.; Shanks, J.V.; Zhao, L.; Gibson, S.I. The ORCA2 transcription factor plays a key role in regulation of the terpenoid indole alkaloid pathway. BMC Plant Biol. 2013, 13, 155. [Google Scholar] [CrossRef] [PubMed]
- Laflamme, P.; St-Pierre, B.; De Luca, V. Molecular and Biochemical Analysis of a Madagascar Periwinkle Root-Specific Minovincinine-19-Hydroxy-O-Acetyltransferase1. Plant Physiol. 2001, 125, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gao, F.; Ren, J.; Lu, X.; Ren, G.; Wang, R. A Novel AP2/ERF Transcription Factor CR1 Regulates the Accumulation of Vindoline and Serpentine in Catharanthus roseus. Front. Plant Sci. 2017, 8, 2082. [Google Scholar] [CrossRef] [PubMed]
- Pani, A.; Mahapatra, R.K. Computational identification of microRNAs and their targets in Catharanthus roseus expressed sequence tags. Genom. Data 2013, 1, 2–6. [Google Scholar] [CrossRef]
- Prakash, P.; Ghosliya, D.; Gupta, V. Identification of conserved and novel microRNAs in Catharanthus roseus by deep sequencing and computational prediction of their potential targets. Gene 2015, 554, 181–195. [Google Scholar] [CrossRef]
- Shen, E.M.; Singh, S.K.; Ghosh, J.S.; Patra, B.; Paul, P.; Yuan, L.; Pattanaik, S. The miRNAome of Catharanthus roseus: Identification, expression analysis, and potential roles of microRNAs in regulation of terpenoid indole alkaloid biosynthesis. Sci. Rep. 2017, 7, 43027. [Google Scholar] [CrossRef]
- Bertaccini, A.; Davis, R.E.; Lee, I.M. In Vitro Micropropagation for Maintenance of Mycoplasma-like Organisms in Infected Plant Tissues. HortScience 1992, 27, 1041–1043. [Google Scholar] [CrossRef]
- Bertaccini, A. Phytoplasma Collection. 2014. Available online: https://www.ipwgnet.org/collection (accessed on 1 April 2023).
- White, J.L.; Kaper, J.M. A simple method for detection of viral satellite RNAs in small plant tissue samples. J. Virol. Methods 1989, 23, 83–93. [Google Scholar] [CrossRef]
- Czotter, N.; Molnár, J.; Pesti, R.; Demián, E.; Baráth, D.; Varga, T.; Várallyay, É. Use of siRNAs for Diagnosis of Viruses Associated to Woody Plants in Nurseries and Stock Collections. Methods Mol. Biol. 2018, 1746, 115–130. [Google Scholar] [PubMed]
- Kellner, F.; Kim, J.; Clavijo, B.J.; Hamilton, J.P.; Childs, K.L.; Vaillancourt, B.; Cepela, J.; Habermann, M.; Steuernagel, B.; Clissold, L.; et al. Genome-guided investigation of plant natural product biosynthesis. Plant J. 2015, 82, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhang, J.; Ewing, A.; Miller, S.A.; Jancso Radek, A.; Shevchenko, D.V.; Tsukerman, K.; Walunas, T.; Lapidus, A.; Campbell, J.W.; et al. Living with genome instability: The adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 2006, 188, 3682–3696. [Google Scholar] [CrossRef] [PubMed]
- She, J.; Yan, H.; Yang, J.; Xu, W.; Su, Z. croFGD: Catharanthus roseus Functional Genomics Database. Front. Genet 2019, 10, 238. [Google Scholar] [CrossRef]
- Kolde, R. Package ‘Pheatmap’. 2012. Available online: https://cran.r-project.org/web/packages/pheatmap (accessed on 1 April 2023).
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Várallyay, E.; Burgyán, J.; Havelda, Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 2008, 3, 190–196. [Google Scholar] [CrossRef]
- Zambon, Y.; Contaldo, N.; Laurita, R.; Várallyay, E.; Canel, A.; Gherardi, M.; Colombo, V.; Bertaccini, A. Plasma activated water triggers plant defence responses. Sci. Rep. 2020, 10, 19211. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, J.; Zhao, S.J.; Hu, Z.B. An active Catharanthus roseus desacetoxyvindoline-4-hydroxylase-like gene and its transcriptional regulatory profile. Bot. Stud. 2014, 55, 29. [Google Scholar] [CrossRef]
- Peebles, C.A.; Hughes, E.H.; Shanks, J.V.; San, K.Y. Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metab. Eng. 2009, 11, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Alonso-Peral, M.; Wong, G.; Wang, M.B.; Millar, A.A. Ubiquitous miR159 repression of MYB33/65 in Arabidopsis rosettes is robust and is not perturbed by a wide range of stresses. BMC Plant Biol. 2016, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant. 2011, 143, 1–9. [Google Scholar] [CrossRef]
- Su, Y.T.; Chen, J.C.; Lin, C.P. Phytoplasma-induced floral abnormalities in Catharanthus roseus are associated with phytoplasma accumulation and transcript repression of floral organ identity genes. Mol. Plant Microbe Interact. 2011, 24, 1502–1512. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.P.; Zhao, H.N.; Zhao, Y.N.; Zhu, B.S.; Yuan, S.S.; Li, S.; Guo, F.Y.; Ji, X.L. MiRNA-seq-based profiles of miRNAs in mulberry phloem sap provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 2018, 8, 812. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef]
- Wu, M.F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef]
- Turchi, L.; Baima, S.; Morelli, G.; Ruberti, I. Interplay of HD-Zip II and III transcription factors in auxin-regulated plant development. J. Exp. Bot. 2015, 66, 5043–5053. [Google Scholar] [CrossRef]
- Li, X.P.; Ma, X.C.; Wang, H.; Zhu, Y.; Liu, X.X.; Li, T.T.; Zheng, Y.P.; Zhao, J.Q.; Zhang, J.W.; Huang, Y.Y.; et al. Osa-miR162a fine-tunes rice resistance to Magnaporthe oryzae and Yield. Rice 2020, 13, 38. [Google Scholar] [CrossRef]
- Várallyay, E.; Válóczi, A.; Agyi, A.; Burgyán, J.; Havelda, Z. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. Embo J. 2010, 29, 3507–3519. [Google Scholar] [CrossRef]
- Tang, M.; Bai, X.; Niu, L.J.; Chai, X.; Chen, M.S.; Xu, Z.F. miR172 Regulates both Vegetative and Reproductive Development in the Perennial Woody Plant Jatropha curcas. Plant Cell Physiol. 2018, 59, 2549–2563. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Feng, Z.; Liu, X.; Bian, L.; Xie, H.; Zhang, C.; Mysore, K.S.; Liang, J. MiR393 and miR390 synergistically regulate lateral root growth in rice under different conditions. BMC Plant Biol. 2018, 18, 261. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Yang, F.; Yu, D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010, 62, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.M.; Rodriguez, R.E.; Mecchia, M.A.; Palatnik, J.F. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 2012, 8, e1002419. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, Q.; Zuo, Z.F.; Liu, L. MicroRNA398: A Master Regulator of Plant Development and Stress Responses. Int. J. Mol. Sci. 2022, 23, 10803. [Google Scholar] [CrossRef]





| miRNA | miRNA | Average RPM of the miRNA | Relative Expression of the miRNA | Cluster | |||
|---|---|---|---|---|---|---|---|
| Vir | WB | NO | Vir/NO | WB/NO | |||
| 156 | ath-miR156a-5p | 77.1 | 33.2 | 46.8 | 1.65 | 0.71 | 5 |
| 157 | ath-miR157a-5p | 503.0 | 569.3 | 523.7 | 0.961 | 1.087 | 1 |
| cro-miR157d-3p | 7.8 | 24.6 | 13.8 | 0.568 | 1.785 | 7 | |
| 159 | cro-miR159a | 5998.2 | 3018.2 | 7460.1 | 0.804 | 0.405 | 4 |
| 162 | cro-miR162a-3p | 111.5 | 121.4 | 184.3 | 0.605 | 0.658 | 6 |
| 165 | cro-miR165a-3p-2 | 87.5 | 118.0 | 163.3 | 0.536 | 0.723 | 8 |
| 166 | cro-miR166b | 589.7 | 455.5 | 392.5 | 1.502 | 1.160 | 3 |
| cro-miR166h-3p | 110,187.1 | 89,050.1 | 110,680.9 | 0.996 | 0.805 | 4 | |
| cro-miR166 | 59,129.1 | 35,752.1 | 49,446.0 | 1.196 | 0.723 | 5 | |
| cro-miR166e-3p-1 | 1133.9 | 1064.7 | 1105.3 | 1.026 | 0.963 | 5 | |
| 167 | ath-miR167d | 26.2 | 26.4 | 49.7 | 0.527 | 0.532 | 6 |
| ath-miR167a-5p | 15.7 | 29.3 | 40.1 | 0.393 | 0.730 | 8 | |
| 168 | ath-miR168a-5p | 318.1 | 551.3 | 1090.5 | 0.292 | 0.505 | 8 |
| 170 | cro-miR170-5p | 10.4 | 12.0 | 19.4 | 0.534 | 0.615 | 6 |
| 171 | cro-miR171c-5p | 8.9 | 2.2 | 0.4 | 21.486 | 5.307 | 3 |
| 172 | cro-miR172g-3p | 19.1 | 29.0 | 6.8 | 2.823 | 4.271 | 2 |
| 319 | cro-miR319e | 97.7 | 69.0 | 83.3 | 1.172 | 0.827 | 5 |
| cro-miR319a | 51.1 | 18.2 | 38.7 | 1.320 | 0.470 | 5 | |
| 390 | cro-miR390a-5p | 6.3 | 9.2 | 20.4 | 0.310 | 0.450 | 6 |
| cro-miR390a-3p-1 | 3.9 | 15.3 | 27.8 | 0.142 | 0.551 | 8 | |
| cro-miR390a-3p-2 | 3.8 | 14.8 | 27.1 | 0.142 | 0.547 | 8 | |
| 391 | gma-miR391-5p | 11.4 | 25.3 | 27.7 | 0.412 | 0.916 | 8 |
| 395 | cro-miR395a | 12.6 | 1.4 | 0.3 | 40.356 | 4.398 | 3 |
| cro-miR395d | 12.5 | 1.3 | 0.2 | 57.137 | 5.850 | 3 | |
| 396 | cro-miR396e | 4137.7 | 1321.1 | 4099.4 | 1.009 | 0.322 | 4 |
| cro-miR396b-5p | 1287.5 | 599.6 | 1283.4 | 1.003 | 0.467 | 4 | |
| cro-miR396h | 3291.7 | 752.5 | 1779.8 | 1.849 | 0.423 | 5 | |
| 398 | cro-miR398 | 1283.5 | 976.9 | 362.3 | 3.543 | 2.696 | 3 |
| novel | cro-novel-34 | 2117.2 | 2422.3 | 992.7 | 2.133 | 2.440 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contaldo, N.; Zambon, Y.; Galbacs, Z.N.; Miloro, F.; Havelda, Z.; Bertaccini, A.; Varallyay, E. Small RNA Profiling of Aster Yellows Phytoplasma-Infected Catharanthus roseus Plants Showing Different Symptoms. Genes 2023, 14, 1114. https://doi.org/10.3390/genes14051114
Contaldo N, Zambon Y, Galbacs ZN, Miloro F, Havelda Z, Bertaccini A, Varallyay E. Small RNA Profiling of Aster Yellows Phytoplasma-Infected Catharanthus roseus Plants Showing Different Symptoms. Genes. 2023; 14(5):1114. https://doi.org/10.3390/genes14051114
Chicago/Turabian StyleContaldo, Nicoletta, Yuri Zambon, Zsuszanna Nagyne Galbacs, Fabio Miloro, Zoltan Havelda, Assunta Bertaccini, and Eva Varallyay. 2023. "Small RNA Profiling of Aster Yellows Phytoplasma-Infected Catharanthus roseus Plants Showing Different Symptoms" Genes 14, no. 5: 1114. https://doi.org/10.3390/genes14051114





