4.1. Candidate Genes Associated with the Flavonoid Biosynthetic Pathway
The
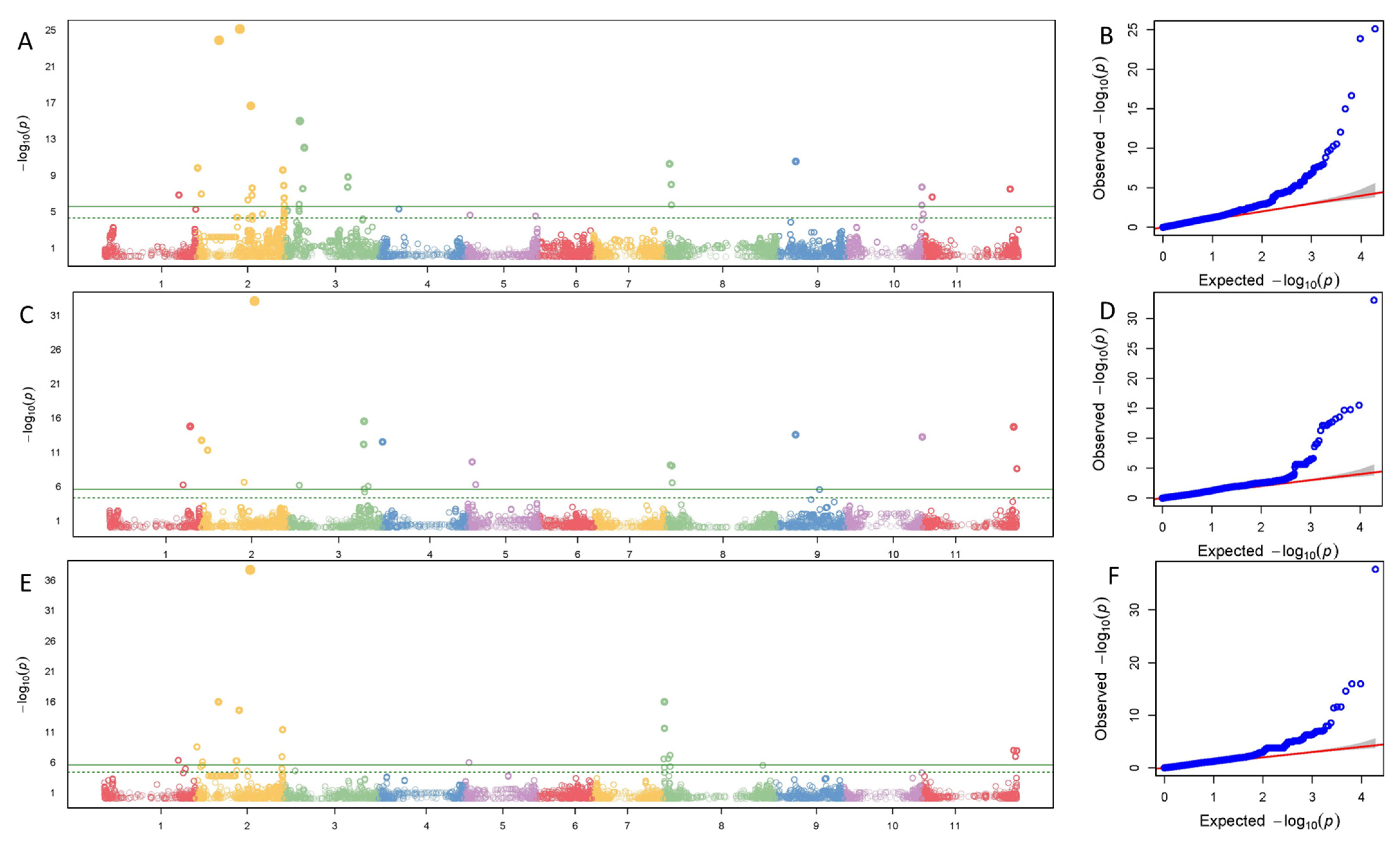
B gene, designated as the precursor of any compound preceding dihydrokaempferol in the flavonoid color pathway, is on Pv02 [
8]. The gene position was located at 48,634,623–48,634,743 bp associated with the SNP ss715645998 [
14] (
Table 1). Three candidate genes (Phvul.002G319500, Phvul.002G319600, and Phvul.002G319800 in G19833 genome) in our study at a distance of about 65–90 kb to an SNP at 48,664,812 bp are involved in the anthocyanin pathway and are potential candidates for
B gene (
Table S1) [
26,
27,
28].
Phvul.002G317000 was found within a 100 kb window of an SNP at 48,416,815 bp associated with C* pod exterior in G19833. It is an R2R3-MYB which have been reported to control anthocyanin biosynthesis in bean tissue and was proposed as a candidate for seed coat color [
29]. This may be a candidate for the red kidney (
Rk) gene and has a sequence approximately 218 kb distance from the
B gene.
Phvul.003G146900 also plays a role in the phenylpropanoid pathway related to seed coat color and was identified as the closest gene to the
Z (zonal partly colored seed coat) locus in bean [
30]. It is within a 100 kb window of SNPs at 35,695,104 bp associated with a*, C*, and H° exterior pod color in G19833 (
Table S1). Phvul.003G203900 was associated with SNPs at 43,010,181–43,092,849 bp for the H° pod exterior in G19833. The gene is also an R2R3-MYB and may regulate color in common bean and cranberry bean seeds [
29,
31] (
Table 6 and
Table S1).
J (formerly dominant
L) was previously identified on Pv10. Researchers determined the location using RAPD markers (OL4S525 and OJ17700, respectively) and these markers were identified as occupying a region of 41,443,673–41,443,694 bp [
4,
20]. We identified a SNP for b* (pod exterior) at 41,403,581 bp on Pv10 in the G19833 reference genome and based on this information, Phvul.010G132400 and Phvul.010G132500 could be candidate genes for
J (
Table S1). These two candidate genes underlie non-darkening seed coat color (
jj) in pinto and cranberry beans [
32,
33]. Phvul.010G132400 encodes for an SNF7 family vacuole sorting protein that in the case of
J, could be involved in vacuolar transport of flavonoids.
J has not been previously identified with altering colors in plant parts other than seeds. In addition to inducing a non-darkening phenotype in seeds,
jj also diminishes flavonol and anthocyanin-based expression. It might have the same effect on these compounds in vegetative tissues, but the effect may be subtle. We were unable to phenotype seeds of the SnAP for
J because the white seed conditioned by
p is epistatic to
J and most accessions in the population are white seeded.
4.2. Candidate Genes Associated with Photosynthetic Pathways
Y gene conferring yellow (wax) pod color has been mapped to Pv02 [
34] (
Table 1). Phvul.002G004400 and Phvul.002G006200 have been proposed as candidates for
y [
15,
16]. The former encodes a pentatricopeptide repeat protein and the latter the SUF family Fe-S cluster assembly protein SufD. Pentatricopeptide repeat proteins can affect chloroplast assembly whereas SUF family genes may have a role in chlorophyll synthesis. Phvul.002G004400 was not found within a 100 kb window in both reference genomes in our study, however, in the former study [
15], the candidate gene search was conducted in a 350 kb window, and if our window is expanded to this distance, this gene could be considered as a candidate for gene y. Phvul.002G006200 associated with H° pod exterior was found in the 5-593 reference genome as Pv5-593.02G005900 (SNP position = 1,468,533 bp,
Table S1). Twelve other candidate genes in the region (Phvul.002G005600–Phvul.002G007100) were considered as possible candidates for
y identified by Yang et al. [
16] before ultimately selecting Phvul.002G006200. These corresponded to candidates Pv5-593.02G005300–Pv5-593.02G006800 found in the 5-593 reference genome. Additional possible candidates identified in the 200 K window bracketing the SNPs (1,620,607–1,631,424 bp) of interest were Phvul.002G014700 and Phvul.002G014800. These were candidates for b* (blue-yellow color) exterior pod color in G19833. These may actually be a single gene model that encodes an Isoflavone 2′-hydroxylase that produces a yellow pigment in the plants [
35].
Phvul.002G007200 (Pv5-593.02G006900) is another potential candidate for
y which encodes for a peptidase protein and has a hydrolase function (
Table S1). Chlorophyllase, which possibly plays a role in chlorophyll degradation during photosystem II (PSII) associated turnover and fruit ripening (but not senescence), is a common hydrolase enzyme in plants.
Chlase or CLH is located in developing chloroplast rather than in mature ones and it helps to protect the leaves from photodamage where chlorophyll turnover is necessary for PSII repair [
36]. Chlase was observed in both green and non-green tissues, but its activity was higher in non-green tissues than in green tissues [
37].
The stay-green trait found in many plant species is classified into cosmetic and functional types. Plants with the functional stay-green genes possess a photosynthesis period longer than normal, while plants with the cosmetic stay green gene remain green but lose their photosynthetic capacity during senescence. Cosmetic stay-green genes play a role in the chlorophyll catabolic pathway [
38,
39]. The cosmetic stay-green trait in common bean is
persistent color (
pc) and some snap bean cultivars have been bred to express
pc because it imparts a more uniform and attractive pod color. Twenty-six accessions in the SnAP carry
pc. A candidate gene for
pc is Phvul.002G153100 (Pv5-593.02G149800) which colocalized with SNPs at 30,590,620 bp and 31,120,266 bp, respectively. These SNPs were associated with b*, C*, and H° pod exterior (
Table S1). The candidate gene encodes for NYE-1, a Mg-dechelatase controlling the stay-green trait in several plant species. Our previous results based on phenotypic characterization showed that
pc types have pod colors that are significantly different from non-
pc types for C* pod exterior [
25]. Even though it was not significantly different for H° from other green colors,
pc had the highest H° among these cultivars. The common link between C* and H° is that both are calculated using b*.
Pv5-593.02G134200 was associated with an SNP at 29,034,300 bp and encodes a protein curvature thylakoid 1D (CURT1) and plays a role in chlorophyll and photosynthesis (PSII) accumulation during de-etiolation, shaping chloroplast and thylakoid membranes [
40]. Pv5-593.02G134900 was also associated with this SNP and is a kinesin-like protein for actin-based chloroplast movement 1 (KAC1) that is affected by blue light and plays a role in chloroplast photorelocation movement that is important for photosynthesis [
41] (
Table 6 and
Table S1).
Phvul.007G103400 is a candidate associated with a SNP at 11,974,591 bp for L* pod interior in G19833 and it is a homolog of Glyma.20G187000 (
Yl) in
G. max. The
Yl gene regulates green color in plant tissues, with the mutant allele
yl producing chlorophyll degradation, reduced PSII activity, and chloroplast structural changes [
42] (
Table S1).
4.3. Candidate Genes from Other Studies
A GWAS study focused on phenolic content used the Bean CAP Snap Bean Diversity Panel included L*a*b* pod color measurements [
43]. Significant SNPs associated with L* and a* were identified on Pv02, and Pv03, and for b* on Pv05. We found SNPs associated with L*, a*, and H° pod exterior on Pv02 within the 100 kb window in the G19833 and corresponding to the same region in 5-593 (
Table S1). Some significant SNPs for flower color on Pv01, Pv03, and Pv09 were also identified in that study and similar positions were found on the same chromosomes in the present study. One was associated with C* pod interior on Pv01 (48,364,135 bp), one for H° pod exterior on Pv03 (43,092,849 bp), and one for L* and b* pod exterior on Pv09 (12,097,310 bp) in the G19833 reference genome. Two SNPs for b* and C* pod exteriors on Pv03, one for L*, b*, and C* pod exterior on Pv09 in the 5-593 were also found (
Table S1). Also, Phvul.009G069401 and Phvul.009G069500 associated with a SNP at 12,097,310 bp, and Phvul.011G212600 and Phvul.011G212800 associated with a SNP at 53,248,080 bp all encode for NAD(P)-binding Rossmann-fold superfamily protein, which may be candidate genes for total phenolic content associated with L* and b* pod exterior in the G19833 reference genome in our study (
Table S1). In each case, the pair of gene models are adjacent to one another and may actually be a single gene model. The last two candidates were also identified as candidate genes for total seed folate content [
44]. Light intensity and quality, which are necessary for photosynthesis, are very important for both folate and phenolic synthesis. Synthesis of these two compounds increases in young organs when light is sufficient [
45].
Common bean pod color was analyzed in a Spanish Diversity Panel and significant SNPs were identified on every chromosome except Pv01 and Pv05 [
35]. The authors found significant QTLs for L* and a* on Pv02 at similar position ranges (790,825–807,178 bp) where we found significant SNPs for L*, a*, and H° pod exterior and interior, and b* and C* pod interior (
Table S1). Another SNP was identified on Pv06 in the Spanish study, and we found it to be associated with C* interior (G19833) in our study. Two QTLs underlying L* and b* on Pv10 were identified in their study, and associated SNPs were proposed. We found these same SNPs for a* and H° pod interior in the G19833 genome at 5,817,088 bp and H° pod interior in the 5-593 genome in our study for the QTL at 7,701,862 bp. The second QTL at 43,424,808 bp was associated with the H° pod exterior in the G19833 genome in our study. It was also associated with leaf color for L*, b*, C*, and H° (
Table S1). Another SNP in the Spanish study for both the number of seeds per pod and a*was also associated with a* pod exterior in the G19833 and for L* pod interior in the 5-593 genomes, both on Pv08 in our study. The Spanish work also identified one SNP for both pod length and b* on Pv01, and two for pod length and color on Pv02. The gene on Pv01 was associated with the C* pod interior in the G19833 genome at 48,364,135 bp (
Table S1). On Pv02, the first SNP was associated with b* and C* pod exterior in G19833 at 47,727,086 bp and for b*, C*, and H° pod exterior in the 5-593 genome at 48,863,854–48,940,147 bp. The second SNP was associated with C* pod exterior, and a* and H° pod interior and exterior in the G19833 at 49,401,491–49,506,969 bp, and for L* pod interior, C* pod exterior, and a* and H° pod interior and exterior in the 5-593 genome at 51,130,674–51,220,858 bp (
Table S1).
Some candidate genes expressed by various traits with high phenotypic variation were found in the other studies. Phvul.001G177500 (at 43,457,300 bp) for pod exterior L* (G19833) was identified as the gene ethylene signaling (
EIN2) that manipulated color by light regulation in pea [
46], and it was a meta-QTL for Fe and Zn content, leading to light absorption on tissue and change the color, on common bean [
47]. It was also a candidate for marsh spot resistance related to Mn deficiency that causes necrosis in cranberry bean seeds [
48] (
Table S1). Phvul.001G181000 associated with a SNP at 43,965,518 bp for b* (middle leaf) in G19833 was identified as an iron-sulfur enzyme fumarase hydratase (
FUM) gene [
49] (
Table S1). This gene was also identified as Pv5-593.01G180000 at 46,993,076 bp, associated with pod exterior L* in 5-593. Phvul.001G182200 encodes UDP-glucosyl transferase and may be involved in glycosylating flavonoid molecules. It is within a 100 kb window for a SNP at 43,965,518 bp associated with leaf b* (middle) in G19833 [
50]. Phvul.001G220300 (Transducin/WD40 repeat-like superfamily protein) and Phvul.001G229600 (ARM repeat superfamily protein) were candidates for seedcoat development in the darkening stage and expressed in the presence of the
P gene in cranberry beans [
31,
51] (
Table S1), and it is associated with a SNP at 47,592,371 bp for C* pod interior in G19833 and as Pv5-593.01G219100 (at 50,692,153–50,804,580 bp) with H° pod exterior in 5-593.
Phvul.002G007200 (a peptidase M20/M25/M40 family protein at 790,825–807,178 bp) was identified in our study as a candidate for L*, a*, H° exterior and interior, and b* and C* interior pod color in G19833 and Pv5-593.02G006900 (at 1,586,653–1,744,283 bp) for pod exterior and interior (except b* exterior) in 5-593. It was previously identified as a candidate for bean seed shape and size [
52]. Plants with light-colored and small seeds have less photosynthetic activity than plants with dark-colored and large seeds [
53]. Phvul.002G021200 (encoding ASYMMETRIC LEAVES 2-like 1) and Phvul.002G021400 a homeodomain-like superfamily protein), both associated with a SNP at 2,190,426 bp in G19833. The corresponding gene models in the 5-593 genome were Pv5-593.02G020900 and Pv5-593.02G021100 (at 2,786,416–3,035,251 bp) were associated with L*, a*, H° pod interior. The former was previously found to be related to abiotic stress while the latter was related to temperature sensitivity [
54,
55] (
Table S1). Phvul.003G203800 (histone deacetylase 6) is a member of the HDAC gene family that plays a role in plant organ development, senescence, and several biotic and abiotic stress responses [
56], and was a candidate for H° pod exterior in G19833 (at 43,010,181–43,092,849 bp) (
Table 6 and
Table S1). Under stress conditions, both the synthesis of photosynthetic pigments and various phytochemical processes are affected [
57].
Phvul.002G152700 (leucoanthocyanidin dioxygenase) was a candidate at 30,590,620 bp for b* and C* pod exterior in G19833 and it is related to delphinidin synthesis. It was downregulated in purple snap beans and was involved in the proanthocyanidin accumulation in the cranberry seed and pinto beans [
31,
58,
59]. The corresponding candidate in 5-593 was Pv5-593.02G149400 at 31,120,266 bp for C* and H° pod exterior (
Table S1). Phvul.010G132433 was a candidate associated with a SNP at 41,403,581 bp for b* pod exterior and Phvul.002G316900 (at 48,416,815 bp), Phvul.006G209500, Phvul.006G209600, and Phvul.006G209700 (at 30,376,061 bp) were all associated with SNPs for C* pod exterior in the G19833. These are all regulatory genes for anthocyanin biosynthesis [
58].
Phvul.004G031900 and Phvul.004G032000 (at 3,910,100–3,975,292 bp) were associated with chlorosis and variation in maturity caused by a deletion in stop codon [
60] and they were a candidate for C* pod interior and exterior in G19833 (the equivalent being Pv5-593.04G036100 in 5-593) (
Table S1). In chlorotic tissues, as photosynthesis and the amount of chlorophyll decreases, the decrease in chlorophyll accelerates, and a color change occurs in the tissues by affecting various pigment syntheses [
61].
Phvul.007G013400 (related to AP2 11) and Phvul.007G014500 (SNF1 kinase homolog 10) associated with an SNP at 941,928 bp were previously found to affect leaf chlorophyll content [
62], and were associated with leaf L* (lower) in G19833. The equivalent genes in 5-593 were Pv5-593.07G012800 and Pv5-593.07G013800 at 1,296,094 bp and were associated with leaf L* (middle).
Phvul.008G019500 (MEI2-like 4) associated with a SNP at 1,633,220 bp in G19833 was a candidate for L* pod interior and a* pod exterior, and previously was identified as a candidate for flower, pod, and seed development [
35,
63]. Phvul.010G032700 (Sec23/Sec24 protein transport family protein) at 4,552,800–4,753,654 bp was a candidate for a*, b*, and C* pod exterior in G19833 and it was previously identified as being involved in the photosynthetic process as a response to water deficiency [
63,
64,
65] (
Table S1).
Sugars are the main source of energy for plant development and various biological processes. Variation in sugar content in pods and seeds is found in common beans, but its quantity depends on whether it is a snap or dry bean, organ size, and the cultivar.
SWEET proteins are involved in photosynthetic carbon transport out of leaves and between cells throughout the plant [
66]. Higher sugar concentration is produced by a higher rate of photosynthesis, which is associated with more chlorophyll and is expressed as darker green colors [
67]. Seed coat color and sugar content are also related [
5]. After flowering, the glucose and fructose fraction in pods decreases while sucrose content increases. Similarly, glucose and fructose content in seeds decreased but sucrose content did not change [
68]. Phvul.002G300900 at 46,900,118 bp (Pv5-593.02G293900 at 48,119,141–48,156,962 bp), Phvul.004G017400 at 2,047,768 bp, and Phvul.009G134300 at 20,332,723–20,375,268 bp (Pv5-593.09G136600 at 22,054,215–22,086,165 bp) for C* and b* pod exterior, Phvul.006G210800 at 30,376,064 bp for C* pod exterior, Phvul.006G000600 at 281,553 bp for C* pod interior, and Phvul.008G007600 at 706,796–706,932 bp for H° pod exterior was found in our study (
Table 6). These candidates were expressed in the flower, leaf, stem, and pod of the common bean where Phvul.002G300900 was downregulated by CdCl
2 and HgCl
2 while Phvul.004G017400 was upregulated by CdCl
2 and NaCl [
69]. Both are nodulin MtN3 family proteins. Phvul.009G134300 (Pv5-593.09G136600), also a nodulin MtN3 family protein, was identified as conditioning resistance to halo blight which is related to the salicylic acid cycle and photosynthesis because exogenous salicylic acid increases photosynthesis under water deficit which could be related to green color in common bean, and plays a role in seed coat development in cranberry bean [
70,
71,
72].
Mineral content may affect the color of some tissues of the plant. Some candidate genes related to nutrient content were found in common beans [
73], and one of them was associated with color parameters in our study. Phvul.003G001300 (tetratricopeptide repeat (TPR)-like superfamily protein) controls magnesium content and Mg deficiency can cause chlorosis in Turkish bean seed [
74]. It was a candidate for b* pod exterior in G19833. Phvul.004G032300 and Phvul.008G045800 (purple acid phosphatase 23 and vacuolar ATP synthase G3, respectively) were related to phosphatase activating protein that gives dark green and purple color [
75]; Phvul.004G032300 was a candidate for C* pod interior and exterior and Phvul.008G045800 was a candidate for H° pod exterior in G19833. Phvul.004G032300 corresponds to Pv5-593.04G036400 in the 5-593 reference genome. Phvul.009G127900 (NRAMP metal ion transporter 6) was related to several nutrients including Fe
2+, Mn
2+, Cu
2+, and Zn
2+ [
48,
76,
77], and was a candidate for a* and b* pod exterior in G19833. Phvul.009G068400 (MRG family protein) was related to phosphorus use and corresponded to L* and b* pod exterior color [
73].