RNA-Sequencing Analysis Revealed Genes Associated with Sweet Potato (Ipomoea batatas (L.) Lam.) Responses to Stem Rot during Different Infection Stages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. RNA Extraction, Library Preparation for Transcriptome Analysis
2.3. Transcriptome Assembly and Screening of DEGs (Differentially Expressed Genes)
2.4. Gene Functional Annotation and KEGG (Kyoto Encyclopedia of Genes and Genomes) Enrichment Analysis
2.5. qRT-PCR (Real-Time Quantitative PCR) Validation
3. Results
3.1. Phenotypic Analysis of Inoculated Sweet Potato Seedlings
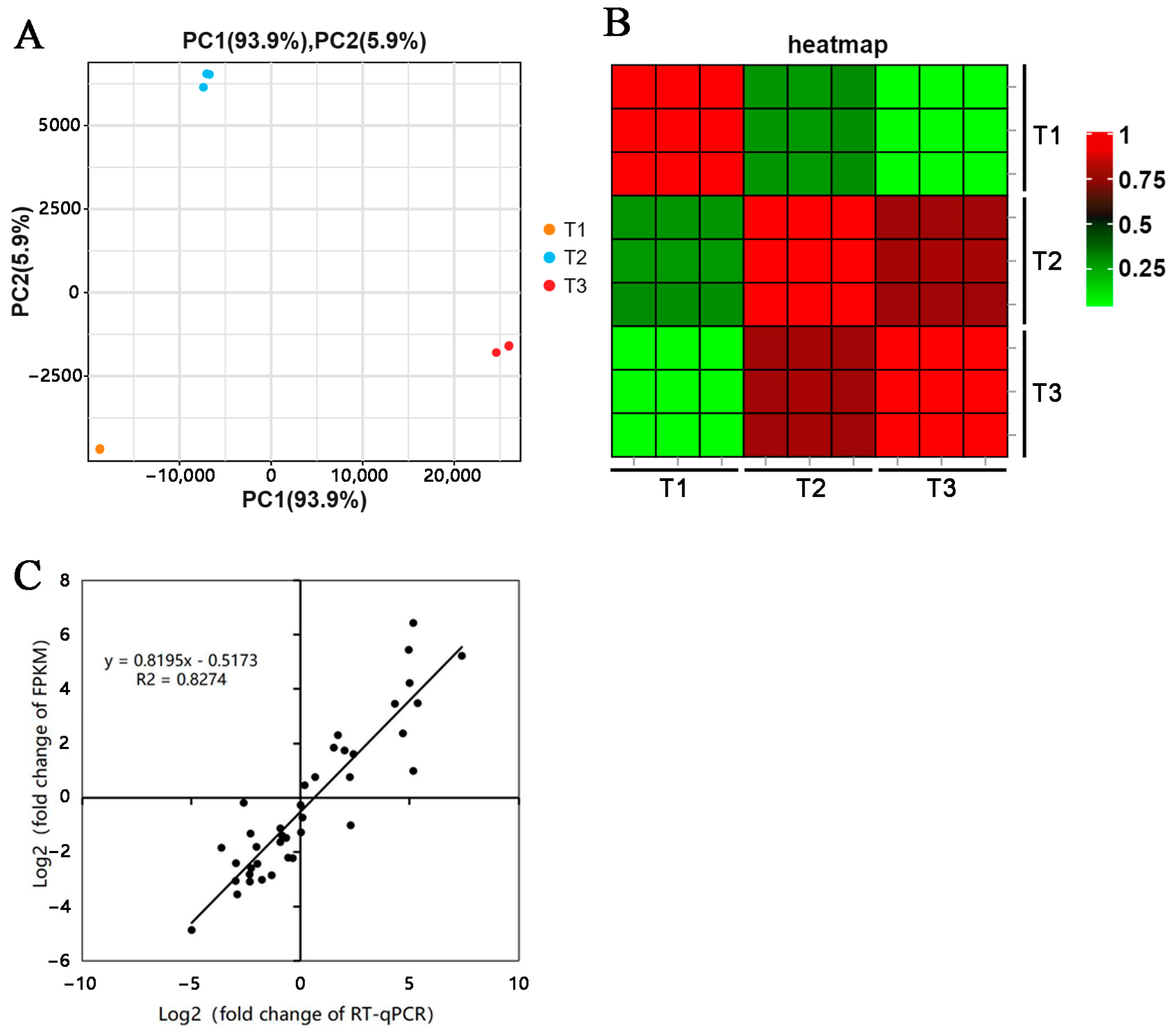
3.2. Transcriptome Analysis
3.3. Identification and Analysis of DEGs
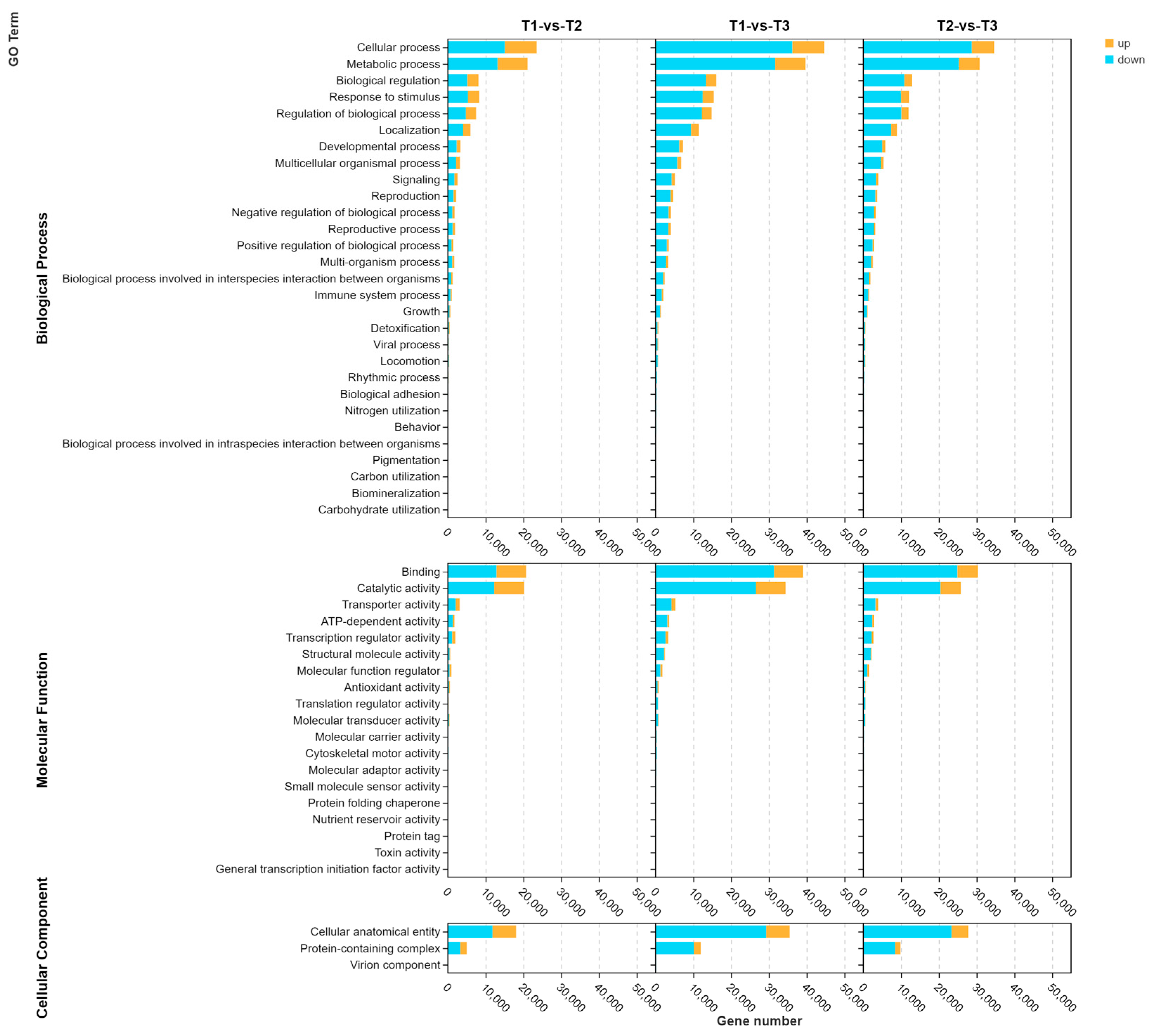
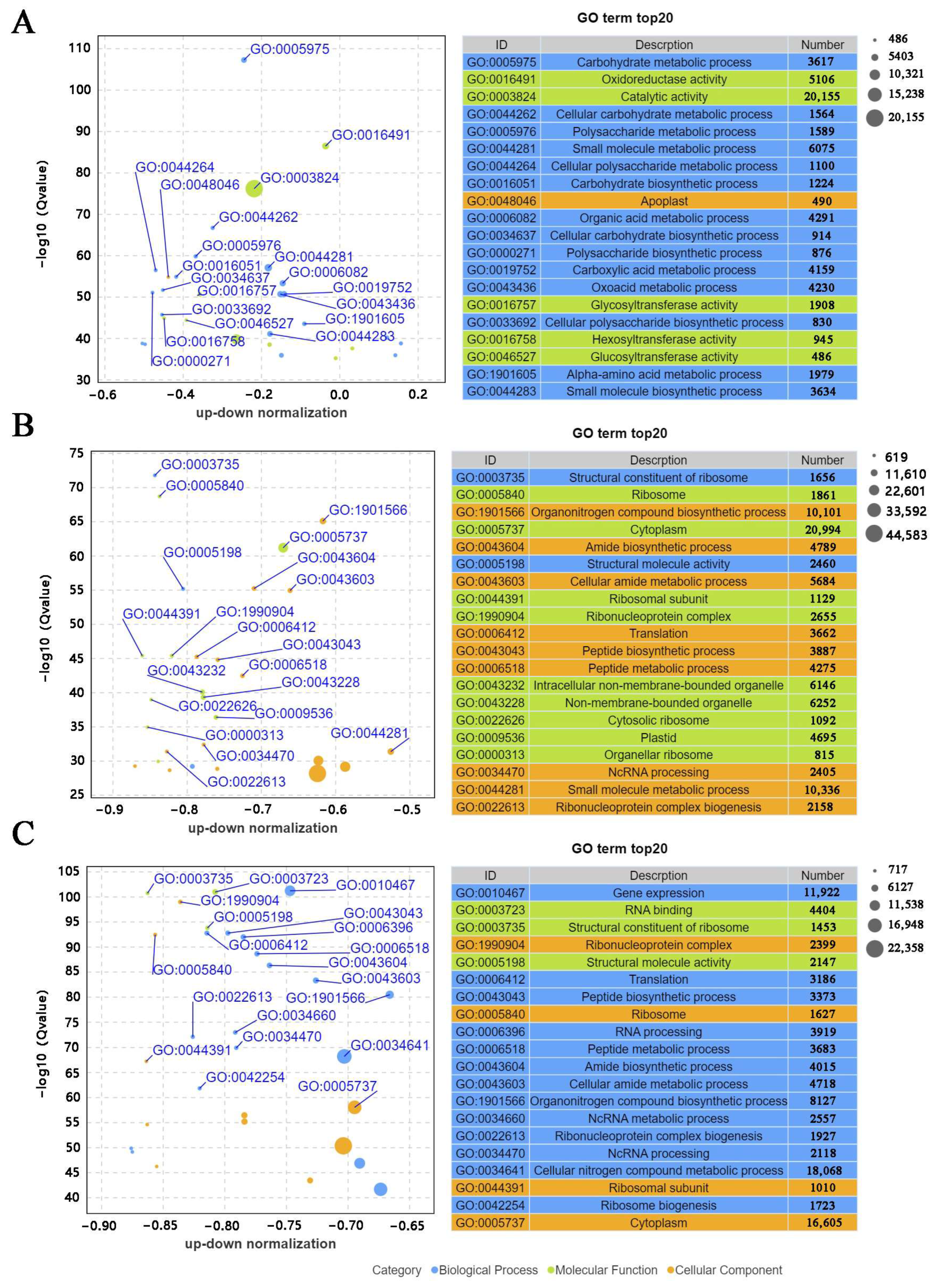
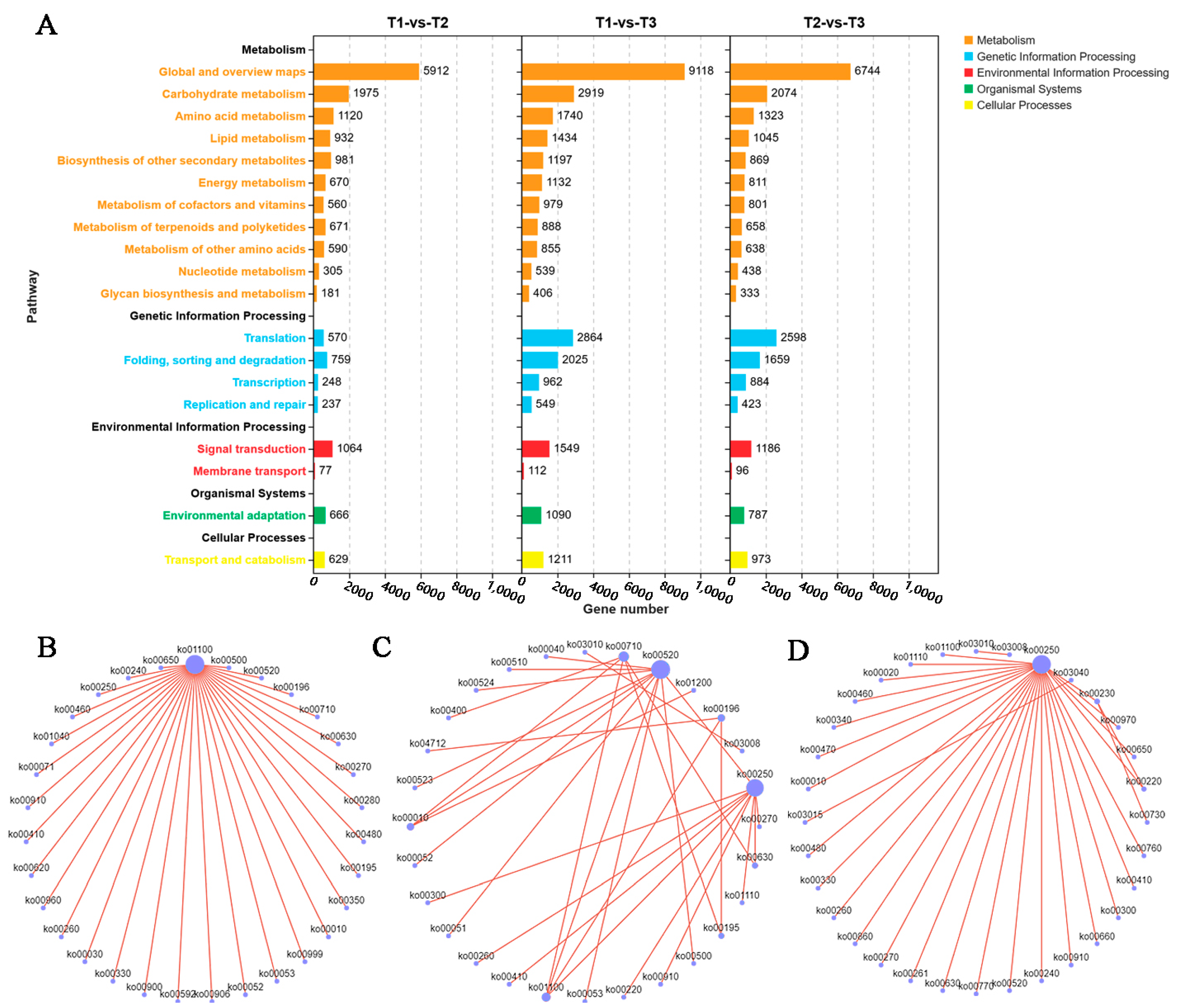
3.4. Enriched GO Terms and KEGG Pathways
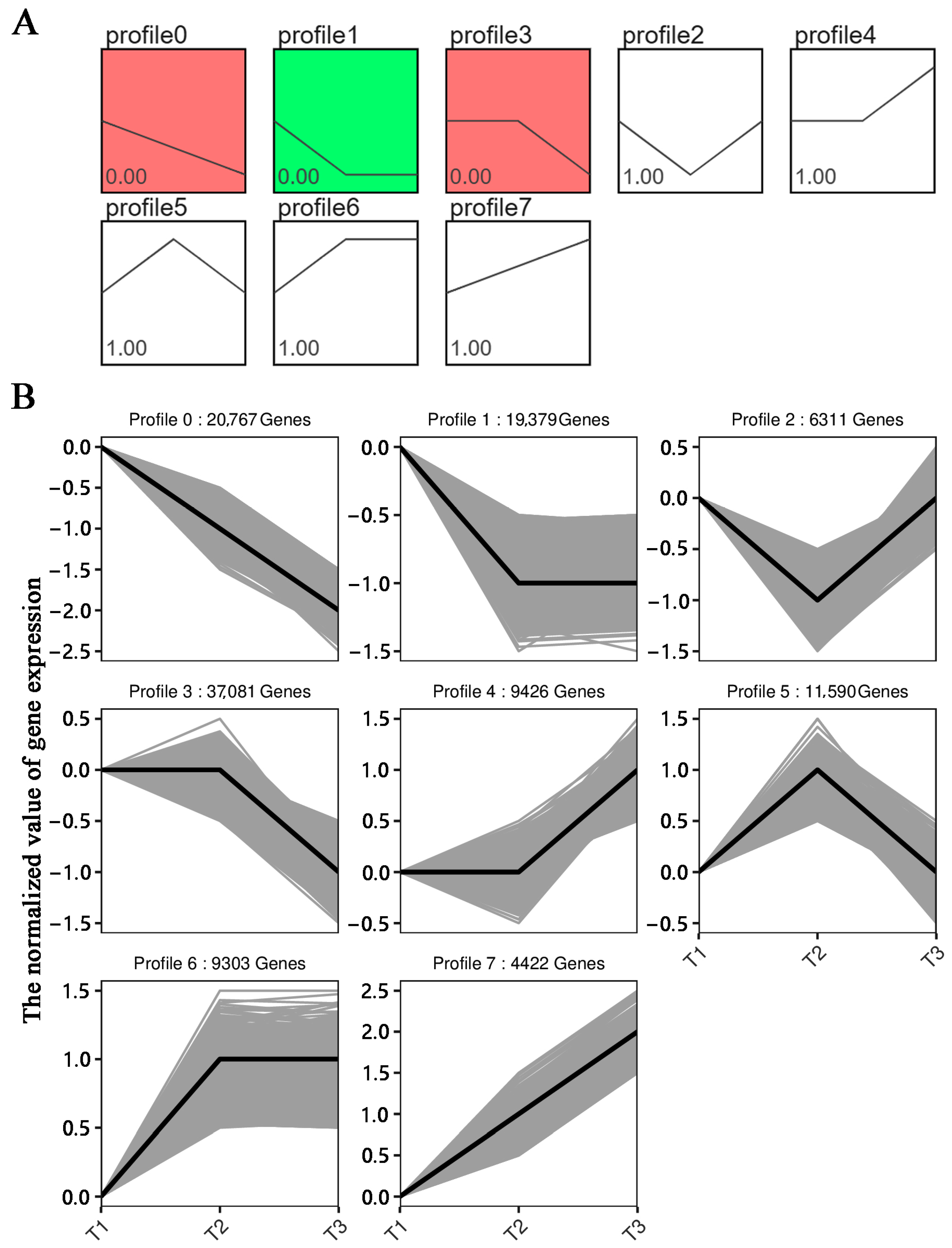
3.5. Analysis of the DEG Expression Trends
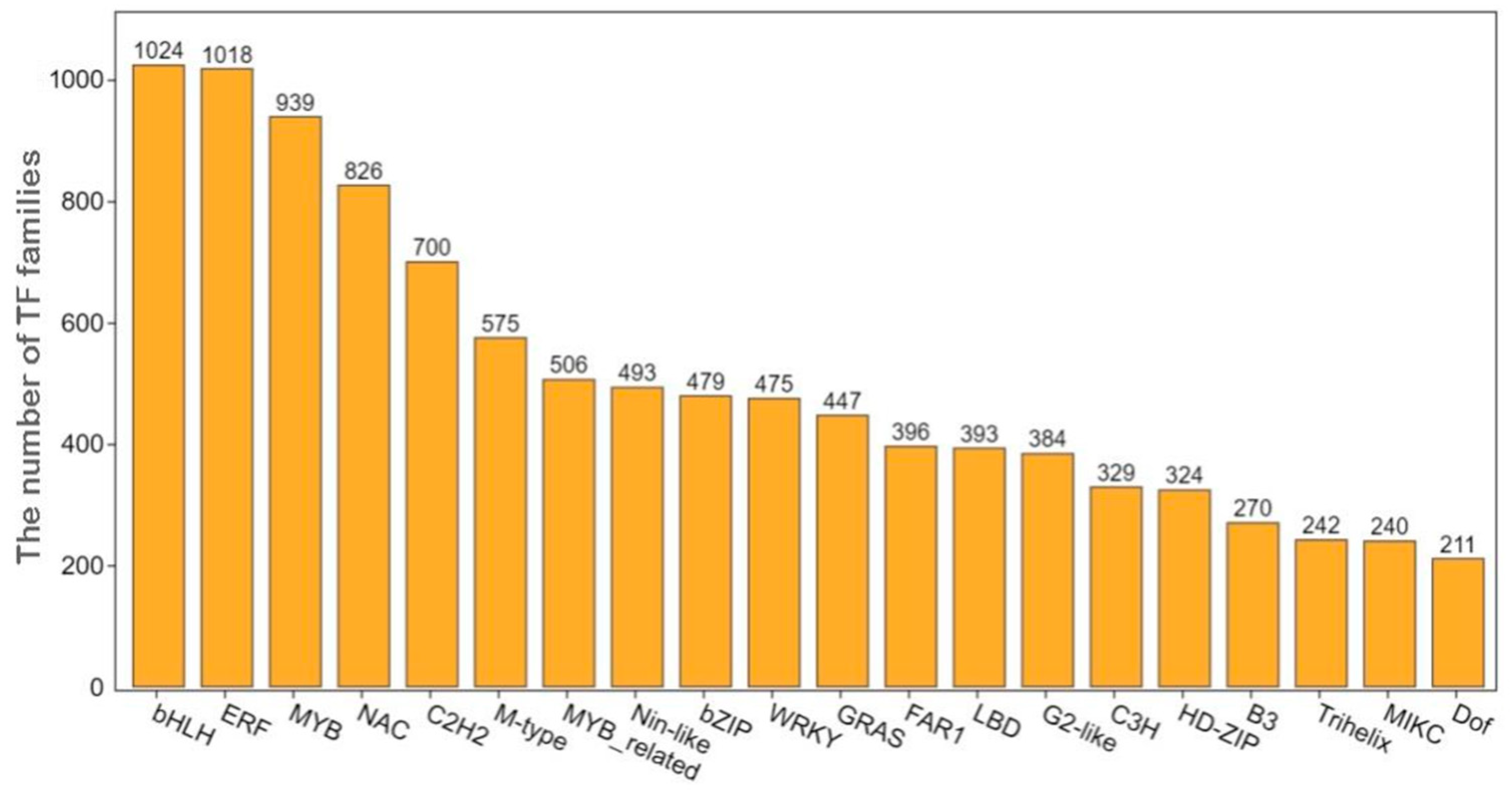
3.6. Analysis of Differentially Expressed Transcription Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Q.; Zhao, H.; Jin, Y.L.; Zhu, J.C.; Ma, D.F. Analysis and perspectives of sweetpotato industry contributing to national food security in china. Jiangsu J. Agric. Sci. 2022, 38, 1484–1491. [Google Scholar]
- Huang, L.F.; Fang, B.P.; Luo, Z.X.; Chen, J.Y.; Zhang, X.J.; Wang, Z.Y. First report of bacterial stem and root rot of sweetpotato caused by a Dickeya sp. (Erwinia chrysanthemum) in China. Plant Dis. 2010, 94, 1503. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.Y.; Huang, L.F.; Ge, C.B.; Liu, Z.J.; Zhang, Y.Y.; Li, J.G.; Fang, B.P. Molecular identification of a new bacterial stem rot disease of sweetpotato in Henan. Crops 2013, 52–55. [Google Scholar] [CrossRef]
- Huang, L.F.; Luo, Z.X.; Fang, B.P.; Chen, J.Y.; Zhang, X.J.; Wang, Z.Y. A new bacterial stem and root rot disease of sweetpotato in Guangdong, China. Acta Phytopathol. Sin. 2011, 41, 18–23. [Google Scholar]
- Schaad, N.W. A Bacterial Wilt and Root Rot of sweetpotato Caused by Erwinia chrysanthemi. Phytopathology 1977, 77, 302. [Google Scholar] [CrossRef]
- Hauben, L.; Moore, E.R.B.; Vauterin, L.; Steenackers, M.; Mergaert, J.; Verdonck, L.; Swings, J. Phylogenetic Position of Phytopathogens within the Enterobacteriaceae. Syst. Appl. Microbiol. 1998, 21, 384–397. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.; Legendre, J.B.; Christen, R.; Saux, M.F.; Achouak, W.; Gardan, L. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov.; Dickeya dianthicola sp. nov.; Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int. J. Syst. Evol. Micr. 2005, 55, 1415. [Google Scholar] [CrossRef]
- Huang, L.F.; Luo, Z.X.; Fang, B.P.; Li, K.M.; Chen, J.Y.; Huang, S.H. Advances in the researches on bacterial stem and root of sweet potato caused by Dickeya dadantii. J. Plant Protec. 2014, 41, 118–122. [Google Scholar] [CrossRef]
- Lou, B.G.; Shen, X.L.; Lu, G.Q.; Lin, X.B.; Li, Y.H.; Zhang, Y.; Lu, Y. Analysis of resistance to stem rot in sweetpotato germplasm resources. J. Zhejiang Agric. Sci. 2018, 59, 2204–2207. [Google Scholar] [CrossRef]
- Ma, B.; Hibbing, M.E.; Kim, H.S.; Reedy, R.M.; Yedidia, I.; Breuer, J.; Breuer, J.; Glasner, J.D.; Perna, N.T.; Kelman, A.; et al. Host range and molecular phylogenies of the soft rot enterobacterial genera pectobacterium and dickeya. Phytopathology 2007, 97, 1150–1163. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Jin, X.; Jia, X.; Wang, H.; Cao, A.; Zhao, W.; Pei, H.; Xue, Z.; He, L.; Chen, Q.; et al. Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genom. 2013, 14, 197. [Google Scholar] [CrossRef] [PubMed]
- De Cremer, K.; Mathys, J.; Vos, C.; Froenicke, L.; Michelmore, R.W.; Cammue, B.P.A.; De Coninck, B. RNAseq-based transcriptome analysis of Lactuca sativa infected by the fungal necrotroph Botrytis cinerea. Plant Cell Environ. 2013, 36, 1992–2007. [Google Scholar] [CrossRef]
- Strauß, T.; van Poecke, R.M.P.; Strauß, A.; Römer, P.; Minsavage, G.V.; Singh, S.; Wolf, C.; Strauß, A.; Kim, S.; Lee, H.; et al. RNA-seq pinpoints aXanthomonas TAL-effector activated resistance gene in a large-crop genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19480–19485. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.L.; Lin, C.; Qian, J.T.; Chou, Z.L.; Chen, J.B.; Sun, C.; Yi, J.P.; Lou, B.G. Characterization of stem and root rot symptoms of sweetpotato and the causal pathogen of the disease. Acta Phytopathol. Sin. 2018, 48, 25–34. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2011, 100, 9440–9445. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Park, S.; Kim, Y.; Ji, C.Y.; Park, S.; Jeong, J.C.; Lee, H.; Kwak, S. Stable Internal Reference Genes for the Normalization of Real-Time PCR in Different Sweetpotato Cultivars Subjected to Abiotic Stress Conditions. PLoS ONE 2012, 7, e51502. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Horvath, D.A.; Staskawicz, B.J. Pivoting the Plant Immune System from Dissection to Deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Wu, S.; He, P.; Shan, L. Phosphorylation of receptor-like cytoplasmic kinases by bacterial flagellin. Plant Signal. Behav. 2010, 5, 598–600. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Nürnberger, T.; Joosten, M.H.A.J. Of PAMPs and Effectors: The Blurred PTI-ETI Dichotomy. Plant Cell 2012, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Schulmeyer, K.H.; Yahr, T.L. Post-transcriptional regulation of type III secretion in plant and animal pathogens. Curr. Opin. Microbiol. 2017, 36, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J.; Jones, J.D.G.; Felix, G.; Boller, T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004, 428, 764–767. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Kracher, B.; Roccaro, M.; Somssich, I.E. Induced Genome-Wide Binding of Three Arabidopsis WRKY Transcription Factors during Early MAMP-Triggered Immunity. Plant Cell 2017, 29, 20–38. [Google Scholar] [CrossRef]
- Li, H.; Sun, J.; Xu, Y.; Jiang, H.; Wu, X.; Li, C. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol. 2007, 65, 655–665. [Google Scholar] [CrossRef]
- Lorenzo, O.; Piqueras, R.; Sanchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 Integrates Signals from Ethylene and Jasmonate Pathways in Plant Defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, H.; Qian, X.; Li, A.; Zhao, G.; Jing, R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef]
- Wang, H.T.; Jiang, J.L.; Wang, Q.; Wei, L.N.; Hu, J. Research progress on the mechanism of R2R3-MYB transcription factors in response to plant stress tolerance. Mol. Plant Breed. 2023, 1–16. Available online: http://kns.cnki.net/kcms/detail/46.1068.S.20230810.1507.012.html (accessed on 17 August 2023).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Zhang, L.; Ji, H.; Song, W.; Zhong, Z.; Jiang, M.; Zhang, Y.; Li, Q.; Cheng, L.; Kou, M. RNA-Sequencing Analysis Revealed Genes Associated with Sweet Potato (Ipomoea batatas (L.) Lam.) Responses to Stem Rot during Different Infection Stages. Genes 2023, 14, 2215. https://doi.org/10.3390/genes14122215
Li C, Zhang L, Ji H, Song W, Zhong Z, Jiang M, Zhang Y, Li Q, Cheng L, Kou M. RNA-Sequencing Analysis Revealed Genes Associated with Sweet Potato (Ipomoea batatas (L.) Lam.) Responses to Stem Rot during Different Infection Stages. Genes. 2023; 14(12):2215. https://doi.org/10.3390/genes14122215
Chicago/Turabian StyleLi, Chen, Liang Zhang, Honghu Ji, Weihan Song, Ziyu Zhong, Meiqiao Jiang, Yungang Zhang, Qiang Li, Linrun Cheng, and Meng Kou. 2023. "RNA-Sequencing Analysis Revealed Genes Associated with Sweet Potato (Ipomoea batatas (L.) Lam.) Responses to Stem Rot during Different Infection Stages" Genes 14, no. 12: 2215. https://doi.org/10.3390/genes14122215




