1. Introduction
Traditional plant-based medicines were once used as the primary source of healthcare, but since the advent of modern pharmaceuticals these preparations have been relegated to the status of ‘complementary’ medicine. In more developed countries, and particularly in the UK, the use of ‘herbal medicines’ has dramatically reduced over the past century. However, recent years have seen the resurgence of biologically active botanicals in many different guises: Food supplements, herbal medicines, nutraceuticals, natural health products and herbal remedies. Each of these types of product have been gaining in popularity, with a recent report showing that the sales of dietary supplements in the US are worth more than ever [
1]. Each different form of product has different requirements in terms of regulation [
2], although it is understood that end users are not necessarily aware of what type of product they are purchasing, and much less what effect that infers on the quality of the product. The booming market for botanicals has been felt in the areas of production; few of the herbals on the market are cultivated and the majority are wild harvested. This has had a drastic effect on the natural populations of some plants, with many medicinal plants listed by the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) [
3,
4] and the number increasing year by year [
5]. The ‘perfect storm’ of high demand, low availability and little regulation is the perfect environment for adulteration and contamination of the supply chain [
4,
6].
As one of the leading traditional herbal medicines in the world, St John’s Wort (
Hypericum perforatum L.) has been the focus of many different techniques for the assessment of quality, efficacy and safety [
7,
8,
9,
10,
11,
12,
13,
14,
15,
16]. This position in the market place also attracted the application of novel, DNA-based, identification techniques as long ago as 2004 [
11]. The identification of plant material using full DNA barcodes has become reasonably standard in research terms, and has even been published as a method within the British Pharmacopoeia for routine use in industry [
17]. However, there are many distinct limitations to the use of full barcode regions for identity:
Time, expense and expertise required for DNA sequencing and analysis.
Inability to detect adulterant/contaminant material.
Limited use with degraded DNA.
Work in this research group initially developed a quick and easy PCR test for the presence of
H. perforatum DNA and showed that this worked well in a small selection of finished products [
13]. The manufacturing process for herbal over-the-counter (OTC) products can damage the DNA of the ‘target’ plant material, meaning that it can become degraded. This was indeed empirically shown by the size of amplifiable DNA products within each extraction; as the amplicon length decreased so the chance of amplifying a product increased. The importance of selecting short ‘mini-codes’ from within larger barcode regions to circumvent this DNA fragmentation was recognised, and the group developed a test resulting in an 80 bp PCR product. This was successfully applied to a sample set consisting of capsules, tables and tinctures [
18].
The development of identification methods in all scientific disciplines is, naturally, a product of the current ‘state-of-the-art’, and as such requires revision over time as methods and technologies develop, and as further information becomes available due to scientific endeavour. Thus, the importance of following an iterative process for test development is imperative, as the authors have previously argued [
19]. The increase in the number of sequences available in public databases for
Hypericum species in the last 10 years has been dramatic, and this extra information needs to be taken into account and incorporated into method maintenance. This has been applied to the assays discussed in this work, redeveloping and applying further checks now that more information is available. The result is an improved technique informed by the current knowledge base and with the added benefit of being a quantitative PCR (qPCR) assay. This development means that the number of starting DNA molecules can be calculated, to a degree. The advantages of using qPCR over barcode sequencing methodologies are numerous, and of particular relevance to industry:
Application to degraded samples due to the dramatically reduced amplicon size.
No requirement to sequence the amplicons.
No requirement to analyse sequencing data.
All results from one machine, and one operator.
While qPCR methods present many advantages over conventional PCR and Sanger sequencing of full barcode regions, it must also be noted that the problem of mixed, adulterated or contaminated samples is not sufficiently covered by a single assay. In this paper we propose the pairing of a species-specific qPCR test with a generic qPCR assay for all amplifiable plant DNA. By providing a measure of ‘total DNA’ and ‘specific DNA’ in a sample, a measure of purity is possible [
20]. However, the robustness and validity of this technique must be proven, and for this purpose we also analyse the sample set using an alternate technique, which is itself a contender for the most powerful method by which to answer the question ‘what is in a mixed plant sample?’: Next Generation Sequencing (NGS).
Since the development of the original PCR test for
H. perforatum, the technology available has changed dramatically. The advent of NGS technologies promises to revolutionise many fields, and the identification of botanicals is no exception. The benefits of this sequencing method over Sanger sequencing are profound, as millions of template molecules are individually sequenced at the same time in a ‘massively parallel’ process. This means that mixed samples can be recognised and the different contributors identified if they have previously been sequenced, and that the detection levels are much lower, giving superior sensitivity. The most well published application of NGS methodologies to botanicals is ‘meta-barcoding’, a method in which ‘barcode’ regions are amplified and all the resulting amplicons are sequenced and identified [
16,
19,
21,
22,
23]. The sequences generated require bioinformatic analysis, and a ‘pipeline’ is designed for this purpose which processes the large data set through several stages: Quality control, where short or low confidence sequences are removed; dereplication, which aims to reduce the effects of PCR bias by selecting only one of each sequence; removal of chimeras, taking out of the equation sequences produced with amplification errors; and finally sequence identification, which is often conducted by searching online databases for matching published sequences, and thereby assigning a species or genus depending on the level of specificity in the results. This process results in a measure of relative abundance for each contributing sequence, which can be displayed in various ways, often a ‘heat map’ [
19]. This is an advanced technique which in many ways represents the current ‘state-of-the-art’, but, as with all methods, it is only ever as reliable as the database to which it refers.
The abundance of available sequence information in public databases is an extreme benefit to the progress of DNA-based identification techniques; however, it does not come without pitfalls, as the number of incorrectly-labelled sequences demonstrates. This situation puts increasing importance on the knowledge and experience of the user, who must apply some criteria for acceptance to sequences acquired from databases before trusting their authenticity and cannot simply depend on the top BLAST result giving a confirmation of species. The use of DNA methods for the identification of a species is entirely dependent on the variation that can be measured between the ‘target’ species, and others that might be present. Therefore, the selection of sequences to differentiate between is a fundamental aspect of assay design. The incorporation of as much information as is available gives the greatest guarantee of specificity and, in order to achieve this, a curated dataset of validated sequences is required. These sequences may well be sourced from online facilities, but curation is essential to confer trust in the resource. The incorporation of sequences from the most closely related ‘sister’ species is the best guarantee of ‘future-proofing’ a method, as it can be deduced that sequences that differ in even the closest relative are likely to differ in all others also. However, as the redesign of this assay shows, the only true certainty can be gained by regular routine maintenance of methods—as is true for all techniques on the bench. For this work, a curated Hypericum database was created and used for the redesign of the qPCR method and also as one of the databases to interrogate with NGS data.
The major benefits of both qPCR and NGS technologies have been described, but of course both methods also have drawbacks. For qPCR, the amount of information generated is much smaller and it is possible that unknown contaminants may not be picked up in the analysis. There is a requirement to standardise each reaction and measure efficiency, as every DNA extraction can behave differently. This has been achieved in several fields [
24] but has not yet been applied to plant methods, which may well be more complex given their complicated genetic inheritance routes. For NGS metabarcoding methods, the effect of amplification biases must again be measured and controlled and this does not yet have a standard approach. Most importantly, metabarcoding is extremely similar to Sanger methods in its requirement for the whole barcode region to be present and not degraded.
Many of these factors have been noted in publications [
21], this is the first to conduct a direct comparison and provide a definitive answer to the question of what is most applicable to industry and regulators, and by doing so to aid in the production of high-quality herbal products for consumers.
2. Materials and Methods
2.1. Sample Collection and Preparation
2.1.1. Commercial Products
A range of different products sold as “St John’s Wort” (SJW) were purchased from high street shops and online suppliers. These included capsules, tablets and tea bags.
Table 1 shows the full inventory of products along with the information provided on the packaging or information leaflets. A number of products carried the Traditional Herbal Registration (THR) logo which conveys a level of quality, whilst similar products were sold as food supplements. Sample number 222 is a mixture of five different plant species, but does not contain
H. perforatum, and was used as a negative control.
2.1.2. Sample Preparation
Non-coated tablets were ground using a pestle and mortar before sampling. Coated tablets were cracked open and sample material taken from the middle to ensure the coating was not included. Capsule halves were separated and the contents emptied into a weighing boat and mixed manually using a spatula before sampling in order to obtain a representative sample. Individual tea bag contents were mixed and then a sample was powdered using a Tissue Lyser (Qiagen, Hilden, Germany). Samples of about 1 g of dried plant material were also powdered using a Tissue Lyser.
2.2. DNA Extraction
DNA extractions were carried out using the Qiagen DNeasy Plant Mini Kit according to the manufacturer’s instructions, starting with 0.02 g powdered plant or commercial product material.
2.3. DNA Quantitation
The concentration of DNA in each extract was determined spectrophotometrically by measuring the A260 of a 2 µL aliquot using a Nanodrop Lite instrument (Thermo Scientific, Waltham, MA, USA).
2.4. Conventional PCR and Gel Electrophoresis
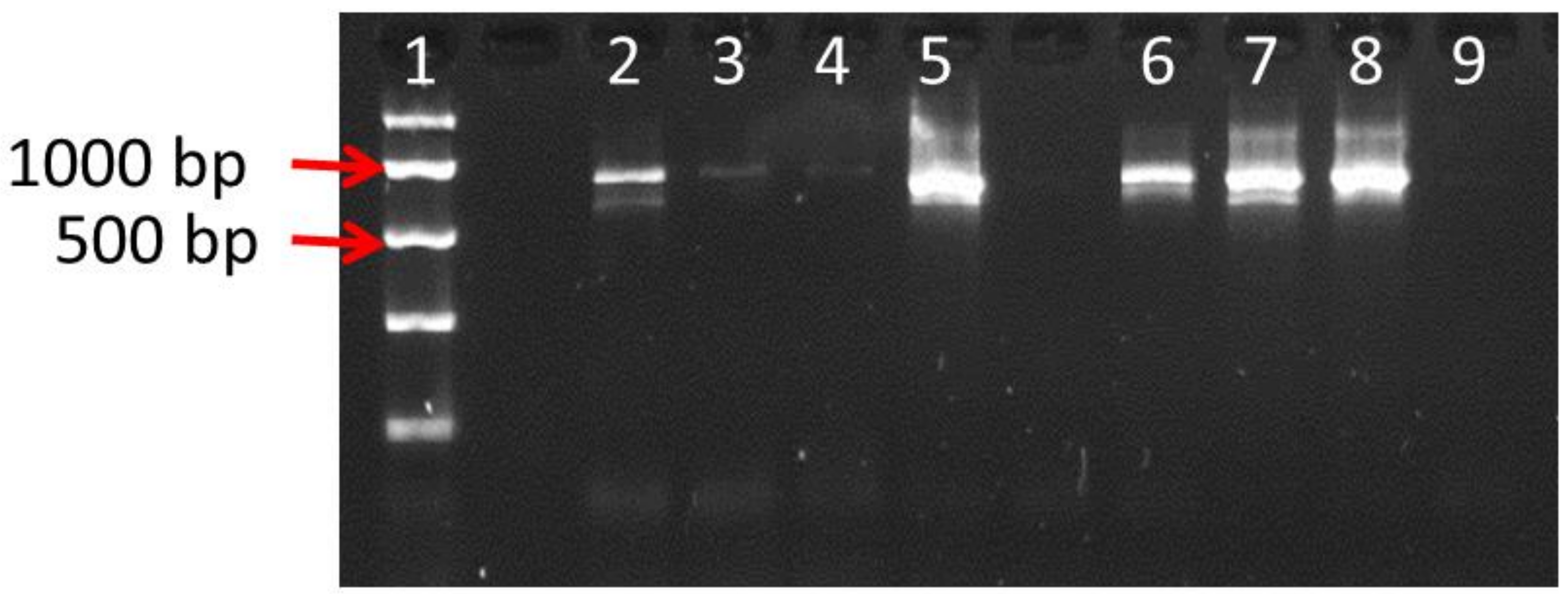
PCR was performed using 1× MyTaq Red Mix (Bioline, London, UK), 0.2 µM of each forward (ITS1 5′-TCCGTAGGTGAACCTGCGG-3′) and reverse (ITS4 5′-TCCTCCGCTTATTGATATGC-3′) primers, and 1 µL of gDNA as template in a total reaction volume of 25 µL. Thermocycling conditions were optimized at 94 °C for 2 min, followed by 30 or 40 cycles of 94 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s, with a final extension step of 72 °C for 2 min. PCR products were run on 2% (w/v) agarose, 1 x TBE gels with 1 µL SYBR® Safe DNA Gel Stain (Invitrogen, Paisley, UK) at 100 V for 30 min and analysed in a Gel Doc™ EZ Gel Documentation System (BioRad, Oxford, UK).
2.5. qPCR Assays with Specific & Generic Primer Pairs
2.5.1. Protocol 1
qPCR assays consisted of EXPRESS SYBR® GreenER™ qPCR SuperMix Universal (1×), relevant primers (0.1 µM each), and template DNA (1 µL) made up to a final volume 20 µL with sterile distilled water (SDW). All reactions were carried out in an MJ Research (Watertown, MA, USA) Chromo 4 real-time thermocycler. The primer pairs used for this assay were: HypGF (5′-CCGTGAACCATCGAGTCTTT-3′) with HypGR (5′-GTCTTACAACCACCGCTGGT-3′) and FO2 (5′-CATAAGAAGTGTAAGGCTCCCGG-3′) used with HRI-S (5′AGAGTCGTTATTGTTATGAACAGAAGGAG-3′). qPCR was performed using three biological replicates with three technical replicates for each sample. Water was run as a negative template control for each test. Thermocycling conditions were optimized at 2 min at 50 °C activation step, 2 min at 95 °C initial denaturation step, 40 cycles consisting of 15 s at 95 °C, 1 min at 60.1 °C, plate read, followed by a melt curve from 54–95 °C at a rate of +1 °C per 10 s, read every 2 °C.
2.5.2. Protocol 2
Each qPCR assays contained 1× Sensifast SYBR green Hi-Rox mix (Bioline), 0.5 µL of gDNA or gBlock, 0.1 µM of each forward and reverse primer in a total volume of 10 µL made up with SDW. qPCR was performed using three biological replicates with three technical replicates for each sample.
Synthetic DNA molecules (gBlocks—IDT, Leuven, Belgium) corresponding to
H. perforatum haplotype 1 (C178),
H. perforatum haplotype 3 (C22),
H. maculatum (C206) and
H. patulum (C203) ITS sequences were constructed as reference standards (
Supplementary Information, Database 1) and dissolved in water to a stock concentration (S) of 10 ng µL
−1. Serial dilutions of the
H. perforatum type 3 standard from S
−3 to S
−7 (10–0.001 pg µL
−1) were performed to generate a standard curve. A working dilution of 0.1 pg µL
−1 of the standard DNA extracts was used as a reference template. Water was run as a negative template control for each test. Thermocycling conditions were optimized at 95 °C for 2 min, followed by 40 cycles of 95 °C for 5 s and 30 s at the primer specific Ta. The melting curve was obtained by heating the amplified template from 65 to 95 °C increasing the temperature by 0.5 °C per cycle.
Primers used in protocol 2 experiments included HypGF, HypGR and FO2 (see Protocol 1) plus 460for (5′-TCG CAA GAG ACA ATC GGG AAT-3′), 460rev (5′-CCA TCC TAT TCC CGA TTG TCT CTT-3′) and 650rev (5′-GTC ACT TTG TGA GTG TTC GAT GTT-3′).
2.5.3. Analysis of qPCR Results
Analyses were conducted according to MIQE guidelines [
25]. Absolute DNA levels were calculated from the C22 synthetic DNA standard curve of Cq plotted against DNA amount. The proportion of
H. perforatum DNA in each sample was determined by calculating the ratio of specific/total DNA using the generic primers as the “reference gene” and compared to the control sample (
H. perforatum C22 dilution S
−5) using the comparative (2
−ΔΔCt) method [
26].
2.6. NGS Methods
Barcode region amplifications were conducted using Hotstar Hifidelity polymerase (Qiagen) as per manufacturer’s instructions, and barcode primers with NGS priming site attachments as follows:
ITS1 region:
Forward: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGAAGKARAAGTCGTAACAAGG-3′
Reverse: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCGTTCAAAGAYTCGATGRTTC-3′
ITS2 region:
Forward: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGATGCGATACTTGGTGTGAAT-3′
Reverse: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACGCTTCTCCAGACTACAAT-3′
After initial barcode region amplification, PCR products were purified (AMPure, Agencourt) and pooled, and a library constructed using these based on the Illumina 16S Metagenomic Sequencing Library Preparation. Index PCR with Nextera XT Index Kit v2 SetC (Illumina, San Diego, CA, USA) was conducted using KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Boston, MA, USA) as per manufacturer’s instructions. The PCR assays were cleaned using AMPure beads following the published protocol, with the following modifications: 1:1 PCR product: AMPure bead initial binding; elution in 10 mM Tris pH 8.0. The resulting libraries were quality controlled using SpectraMax Quickdrop, Qubit HS dsDNA kit and were then run on Bioanalyzer with a HS DNA chip, after being diluted to approximately 1 ng/µl. Using these findings, the libraries were normalised to 4 nM each, and were denatured and diluted following the standard 16S protocol. A 40% PhiX spike-in was included in the sequencing run to compensate for low library complexity, as per the protocol. The complete library was loaded onto a MiSeq v2 500 reagent cartridge and were sequenced 2 × 250 bp paired-end reads.
2.7. Bioinformatics
2.7.1. Curated Database of the ITS Region of Significant Hypericum Species
A small database of ITS sequences from significant
Hypericum species was constructed (
Supplementary Information, Database 2). Species were chosen if they matched at least one of three criteria:
Close relatives of H. perforatum.
Common in commercial trade as ornamental or medicinal plants.
Reported to be found as adulterants of H. perforatum.
ITS sequences from each target
Hypericum species were identified in GenBank and “seed” sequences were chosen from recently published studies based on large collections of vouchered specimens. The seed sequence was used to identify similar sequences using Blast Explorer to produce a crude phylogenetic representation using BLAST scores and an estimate of the final multiple alignment length [
27]. This initial screen allowed clusters of similar conspecific sequences to be identified, and for anomalous conspecific sequences on distant branches of the tree to be excluded from the dataset. Clustered conspecific sequences were aligned and a consensus sequence was constructed by majority voting at each polymorphic position. In small datasets, it was often possible to identify a “type” accession sequence which reflected all of the majority polymorphisms. In large species datasets where it was possible to clearly define different haplotypes, these were recorded separately (e.g., three
H. perforatum types and two
H. maculatum types were recognised). All sequences were trimmed to start at the first base of the ITS1 region and finish at the last base of the ITS2 region.
2.7.2. Next-Generation Sequencing Amplicon Processing
The raw paired-end reads were trimmed and quality filtered using Trim galore! v. 0.3.3 [
28], a wrapper tool around Cutadapt [
29] and FastQC [
30]. The minimum phred quality score required for further processing the reads was set to 28. The surviving reads were then further processed with USEARCH v. 9.2.64 [
31]. Specifically, reads with identical sequences were reduced to a single read while keeping the number of contributing reads in the header. Sequences that were made up from less than 10 reads were discarded. In the next step, chimeric sequences were removed with the
-uchime2_denovo algorithm [
32,
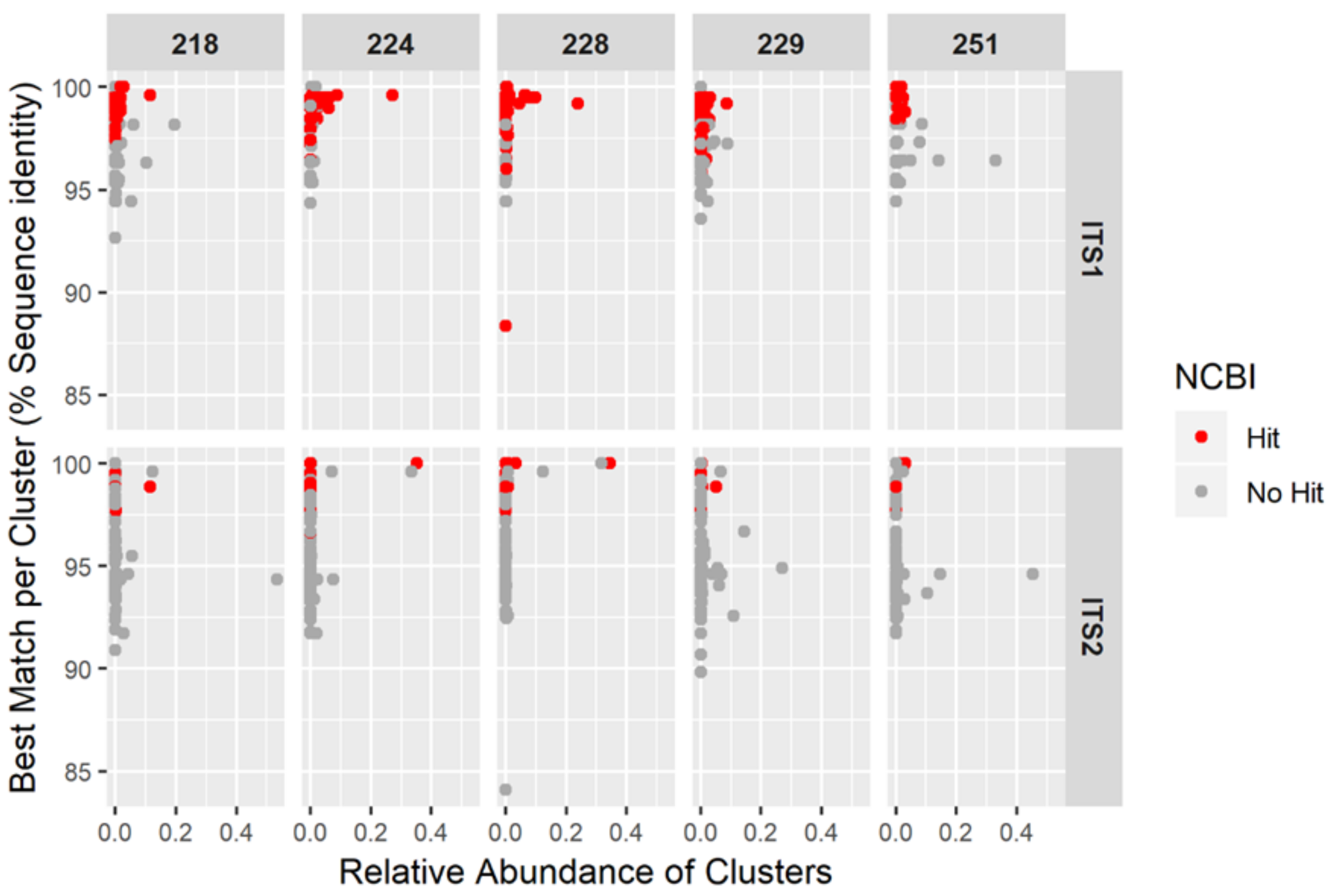
33]. The final steps consisted of summarising sequences with >0.99 identity to a single cluster. The remaining clusters were then separated into two different files, containing either the ITS1 or the ITS2 clusters.
The clusters were then queried to the nucleotide NCBI and the local database using nucleotide blast (blastn-megablast) in BLAST+ v. 2.2.26 [
34]. The local database consisted of the
Hypericum samples described above.
The resulting blast output files were then imported to MEGAN v. 5.11.3 for taxonomic profiling. The lowest common ancestor (LCA) parameters were set to minScore = 50, maxExpected = 0.01, topPercent = 3 and minSupport = 1. The taxonomic information and the number of reads contributing to a specific taxonomic hit was exported to a species-abundance table and further processed with custom R scripts and the R package ‘tidyverse’ [
35]. In the case that the blast search against the NCBI database returns hits to several, potentially distant, taxa, the next deeper taxonomic level is returned. Some of these hits were very unspecific and resulted in very deep taxonomic levels, such as Pentapetalae. In this analysis, such categories were treated as “no hit”. The relative abundance of reads in each cluster was calculated relative to the total number of assigned reads per sample (including the reads treated as “no-hit”). Taxonomic categories that were less abundant than 2% were excluded from further analysis. The filtered dataset, containing the taxonomic categories, together with their relative abundance per sample were displayed in a heatmap. The heatmap was calculated for each region alone and as a combined dataset. In order to compare the NCBI blast results to the local BLAST search, the clusters which resulted in a hit to genus
Hypericum or one of its species in the NCBI search were further investigated. The best matching taxon per cluster was extracted. This was based on the percent sequence identity (“pident”), rather than the e-value or the bit score.
4. Discussion
The application of qPCR and NGS metabarcoding to discern identity and purity of herbal products was assessed using 19 samples from a variety of processed products claiming to contain St John’s Wort. Around half of these products were not suitable for either qPCR or NGS analysis due to the low yield and/or poor quality of DNA extracted from these materials. This is an anticipated finding, and relates directly to the composition of the products under analysis. The samples yielding the highest concentrations of amplifiable DNA are largely made up of plant material that has undergone minimal processing, such as the teas (samples 218 and 229). Conversely, many of the samples displaying low abundance of DNA are produced from extracts, and may well contain no plant material, such as samples 213 and 230—each tablet containing standardized extracts. In this situation the anomalies become of interest also, such as samples 235 and 244 that are labelled as containing a mass of ‘SJW’, but yield no DNA. The meaning of this label statement is not clear, and could equally refer to plant material or extract, and clarity on this may well be useful to the consumer. It is possible that techniques such as ‘baiting’ coupled with NGS would be capable of analysing these materials, but these results generally confirm the long-held view that molecular methods are not suitable for routine quality testing of processed extracts, and are better applied further up the supply chain, ideally focussing on raw materials, as described previously [
19]. However, this study has adopted the workflow of a forensic investigation, using the initial FO2/HRI-S PCR assay as a presumptive test and three investigative tools (ITS barcoding, qPCR and NGS) to analyse those samples in which traces of DNA were detected. It is perfectly legitimate to apply these tools to determine the quality of market products provided that the underlying concepts of forensic investigation are fully understood.
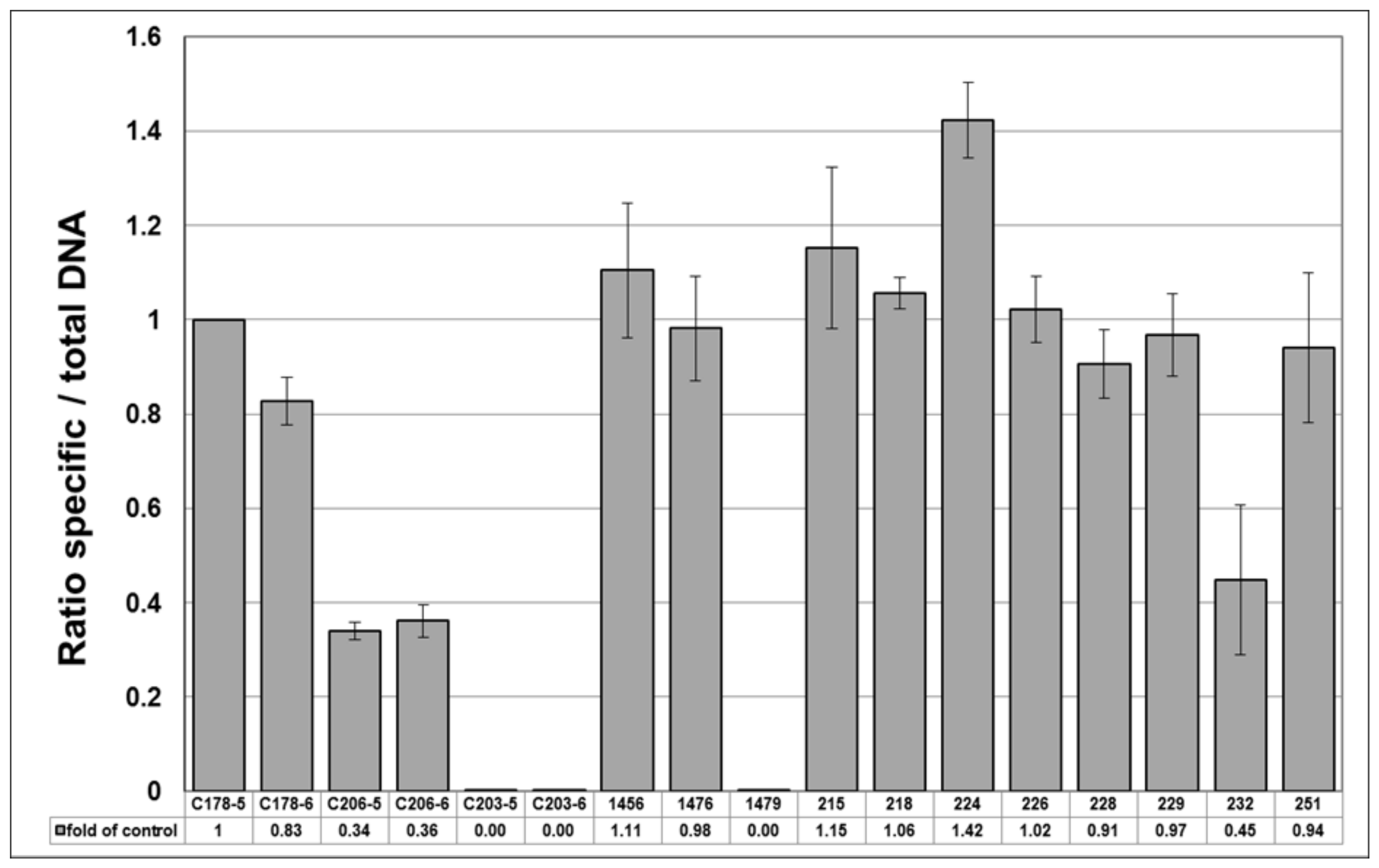
The remaining 10 samples fell into two distinct categories based on whether they produced templates that were suitable for NGS methodologies (
Table 6). It is worth noting that the samples that produced NGS data had significantly more DNA and were less fragmented than those that did not. This suggests that a lower ‘cut-off’ limit of DNA concentration may be applicable to NGS metabarcoding methodologies, and further investigations to show exactly where these boundaries lie would be a useful addition to the field. There is a good correlation between the qPCR and NGS metabarcoding results for the five samples that produced both. They all had qPCR ratios of specific/total
Hypericum DNA >0.90, consistent with the strong
Hypericum signal in the NGS analysis. Samples 218 and 224 in particular had qPCR ratios >1.00 and good quality NGS profiles, consistent with the presence of good quality
H. perforatum DNA.
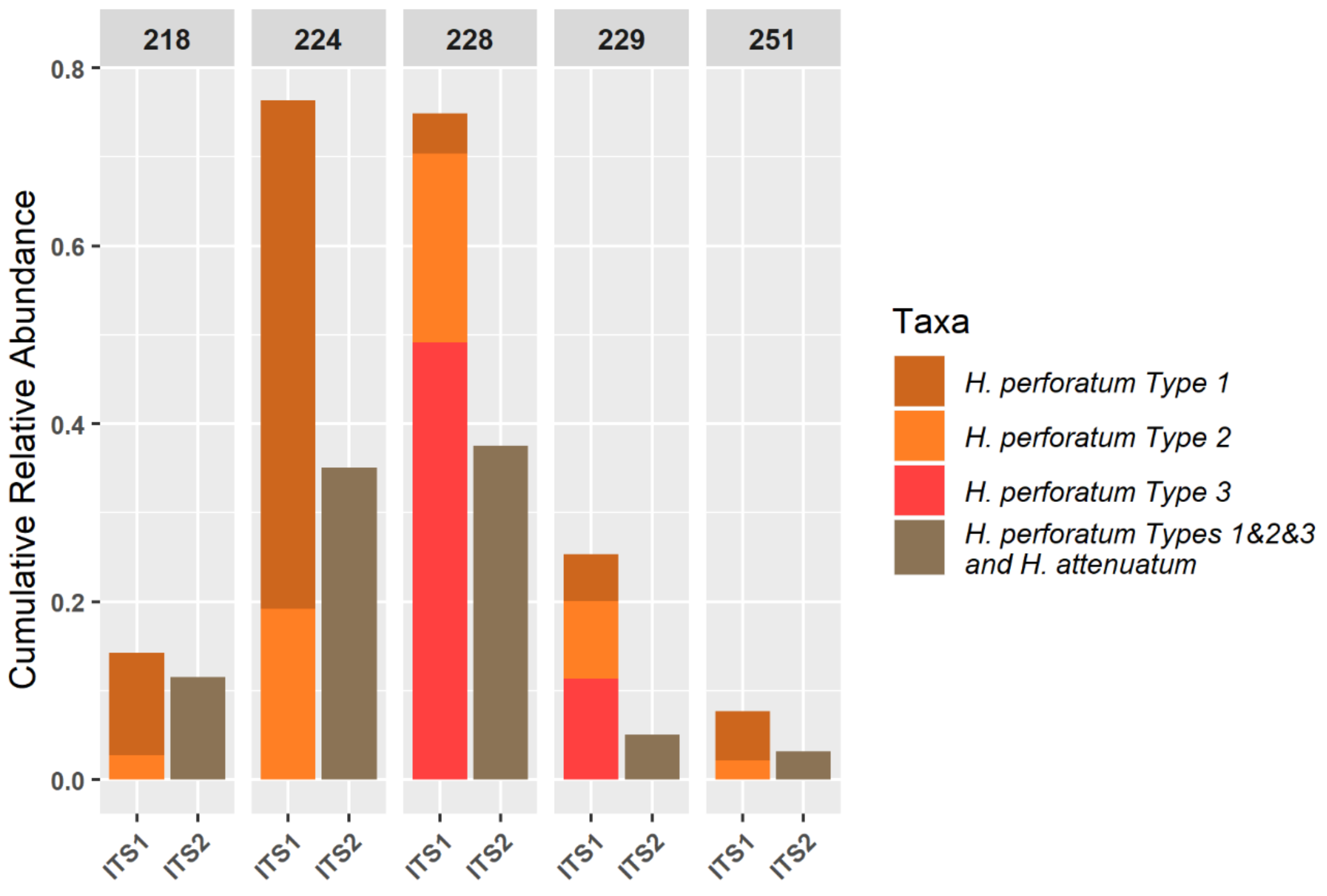
The qPCR results of samples 229 and 251 show ratios of 0.97 and 0.94, respectively (
Table 6), which could indicate low levels of contamination by other
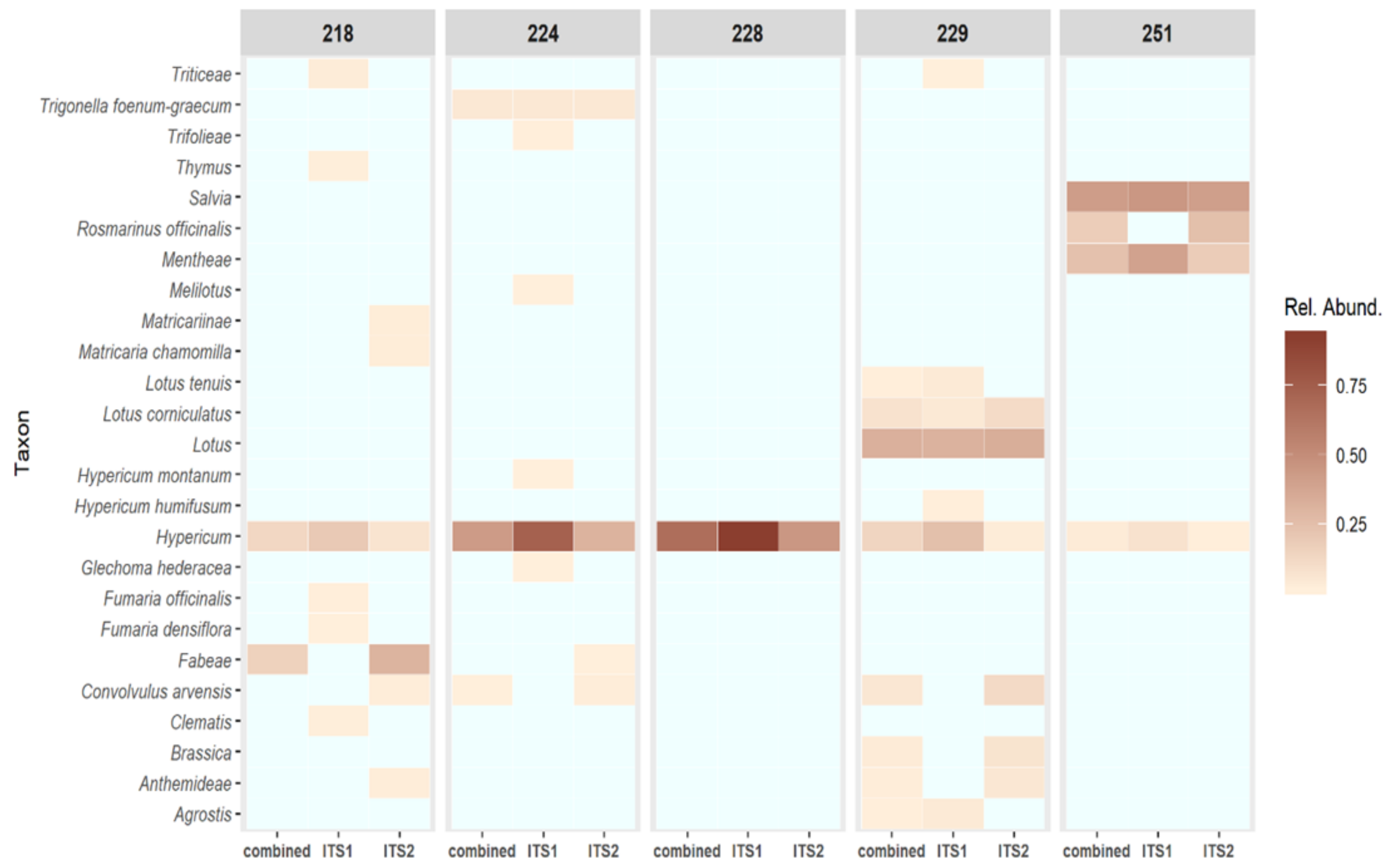
Hypericum species. This is consistent with the NGS profiles, which show some contamination of both samples by a range of commercial herbal plant species (
Figure 4) and a low relative abundance of identified
H. perforatum sequences (
Figure 6). It is likely that this is the result of cross-over in manufacturing processes, given the types of species present. It should be noted that the HypG primers are universal for ITS sequences of the
Hypericum genus but may well not amplify and therefore measure the DNA that is picked up as background in the NGS method. The use of total plant generic primers targeting the conserved 5.8S region [
39] would be a useful addition to this work.
It has previously been claimed that NGS metabarcoding methods are too sensitive for herbal samples, which are by nature likely to contain low-level contamination below an acceptable threshold. However, using a logarithmic proportional scale displayed via a heatmap, this study has shown that even a finished product can give an ideal ‘pure’ result. However, more work is required to explore where the boundary of ‘acceptable’ should lie with respect to contamination. The novel use of a smaller, curated database to further profile sequences within the target genus yielded high specificity—showing the different types of
H. perforatum present in each sample (
Figure 6). The resolution of
H. perforatum genotypes could be used to investigate the genetic basis of phytochemical variation in
H. perforatum and would be extremely useful to companies for identifying and maintaining the quality of their sourced plant materials.
The laboratory methods and bioinformatics pipeline required to complete this analysis is significantly more demanding on the analyst than the qPCR method. This remains a barrier to entry for new users into this field and can prevent even university-based research groups from accessing NGS technology. As time and methods progress, this is likely to become less of an issue, but the added benefit of these methods for herbal drugs must be compelling in order to persuade industry to invest.
The approach used in both the original simple PCR test for H. perforatum and this improved and quantitative qPCR method represents an accessible route to entry into DNA quality control testing for industry with lower capital costs and, more importantly, less analytical skill required compared to NGS metabarcoding. The results also indicate that qPCR is more resilient to fragmented DNA samples than metabarcoding, as is to be expected from the nature of the methodologies, as the qPCR method utilises shorter fragments. However, even with the qPCR method less than half of the samples produced convincing results.
NGS metabarcoding provides a significantly higher level of detail about the constituents of a DNA sample, giving information not only on the presence of ‘target’ species (Hypericum in this case) but also broadening the scope and identifying un-anticipated contaminants. This study has shown the utility of this, where the ‘panel’ of adulterants and contaminants was made up of Hypericum species, and therefore the qPCR test did not directly pick up contamination with distantly related herbal plants. The resolution offered by NGS metabarcoding enables the monitoring of supply chains and production methods and can provide clues as to the potential sources of contamination, for example crossing over from production equipment as discussed earlier.
While there is debate around the application of molecular methods toward quantitative information on plant materials, qPCR methods have been applied in this respect to many other areas such as GMOs, food and viral load. The question of this application relates back to the point in the supply chain where sampling occurs and would be achievable when analysing raw materials. For this to be validated, standards would be required for each qPCR, as the DNA matrix can have an effect on the reaction, which would confound results if unmonitored. The standards used to calibrate this method could be further adapted to provide such a reference, allowing standardisation of the technique. The question of quantitative validation is more problematic in the NGS metabarcoding scenario due to its broad scope, which is also its major benefit. If all species are the ‘target’ for the method, then considerable sequence variation can occur between ‘universal’ priming sites. This means that problems of preferential amplification are likely to occur, and, for example, shorter ITS regions, such as those in fungal genomes, could become misleadingly abundant due to the propensities of methods required for library construction.
Many studies have recently underlined the need for better quality control and traceability of herbal products in the chain from field to food in all the continents [
19,
22,
40,
41,
42,
43]. DNA-based testing using qualitative PCR is already the prevailing method used for the identification of foodborne pathogens and for assuring food safety and, as recently suggested by Newmaster et al. (2019), this method delivers precise, sensitive, specific and reliable results. Newmaster et al. (2019) recommend that quantitative PCR DNA-based assays should be used to test plant material available on the market throughout the supply chain to verify and quantify possible adulterations and contaminations. Recent studies have also proved that DNA metabarcoding is a valid qualitative analytical approach to investigating discrepancies between products and products’ labels in complex multi-ingredient and processed mixtures [
44].
The more reductionist a technique, the more vulnerable it is to the acquisition of new information. For this reason, the most applicable industry methods, with the most simple and easy approach must be iterative and change over time. Maintenance is key to preserving the integrity of any assay, allowing for previously unsampled natural variation and investigation of ‘Out of Spec’ (OOS) results. These findings must then be fed back into the system, generating a manageable maintenance routine for analysis. Weighing up the benefits and limitations of the two techniques investigated, the most efficient proposal for this is to employ routine qPCR techniques coupled with NGS metabarcoding for the understanding of OOS results. This would enable the redesign of the qPCR based on the known contaminants, as found over time using NGS metabarcoding.
The power and utility of DNA-based methods for the identification of herbal drugs is in its direct measurement of species, whereas other methods focus on secondary processes within the plant which are not fixed. However, the most efficient use of this information may be in increasing the stringency of other analytical methods. By designing high quality phytochemical or physical testing methods around materials that have been identified using DNA-based methods, the boundaries for ‘pass’ or ‘fail’ can be made more rigorous and can ensure that all unacceptable variation is ruled out. The inclusion of all methods and information for the design of quality control methods is fundamental; only by analysing all features can the most important and measurable discerning features be understood, and for medicinal plants molecular identity is an essential part of this picture.