ScDREB10, an A-5c type of DREB Gene of the Desert Moss Syntrichia caninervis, Confers Osmotic and Salt Tolerances to Arabidopsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. DNA/Protein Sequence and Phylogenetic Analyses
2.3. Subcellular Localization Analysis
2.4. Analysis of Transactivation Activity of ScDREB10
2.5. Generation of ScDREB10-Overexpressing Arabidopsis
2.6. Evaluation of the Osmotic and Salt Stress Tolerances of Transgenic Arabidopsis at Germination Stage
2.7. Evaluation of Osmotic and Salt Stress Tolerances of Transgenic Arabidopsis at Seedling Stage
2.8. Determination of the Potential Down-Stream Genes of ScDREB10
2.9. Statistical Analysis
3. Results
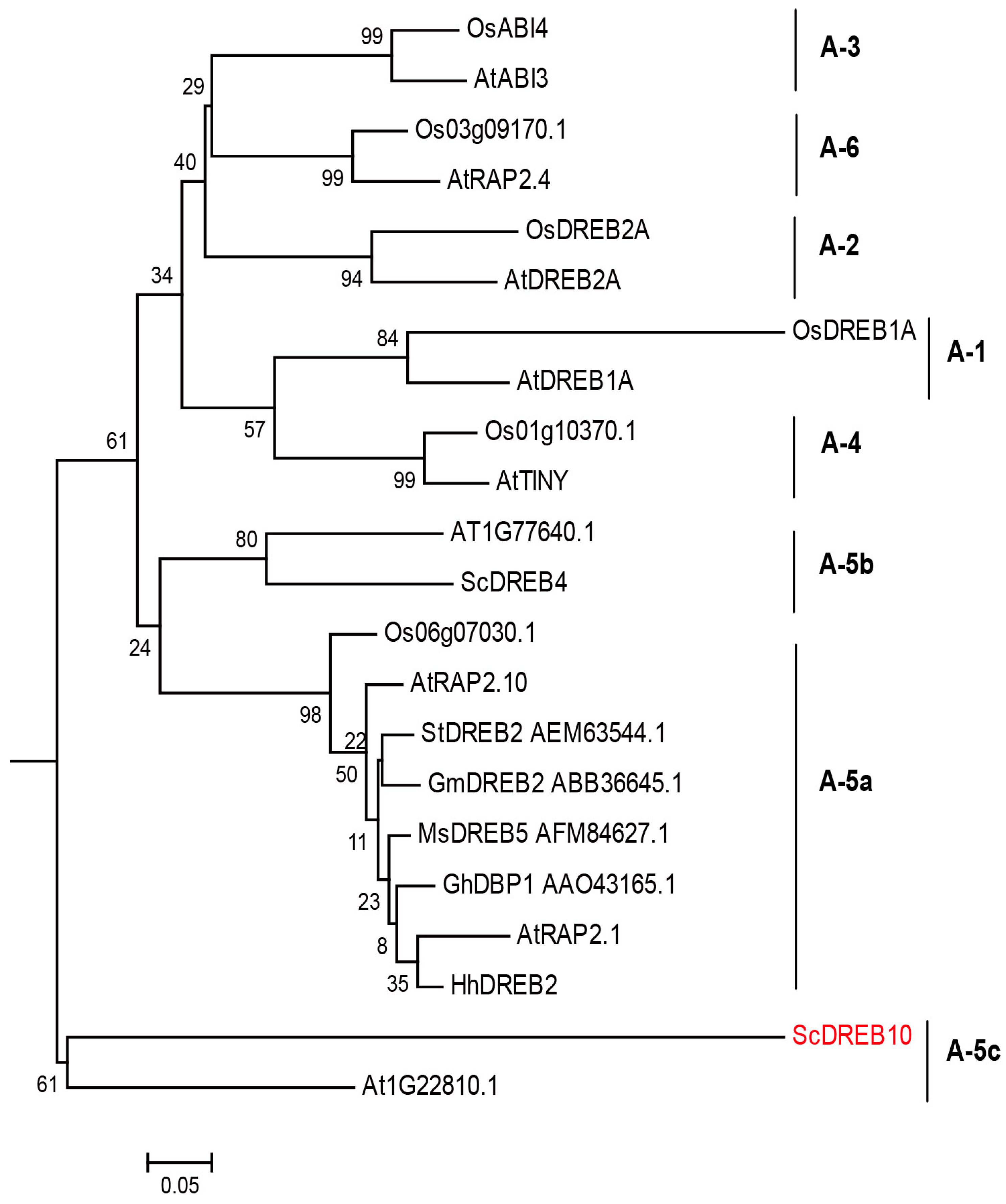
3.1. Phylogenetic Analysis of ScDREB10
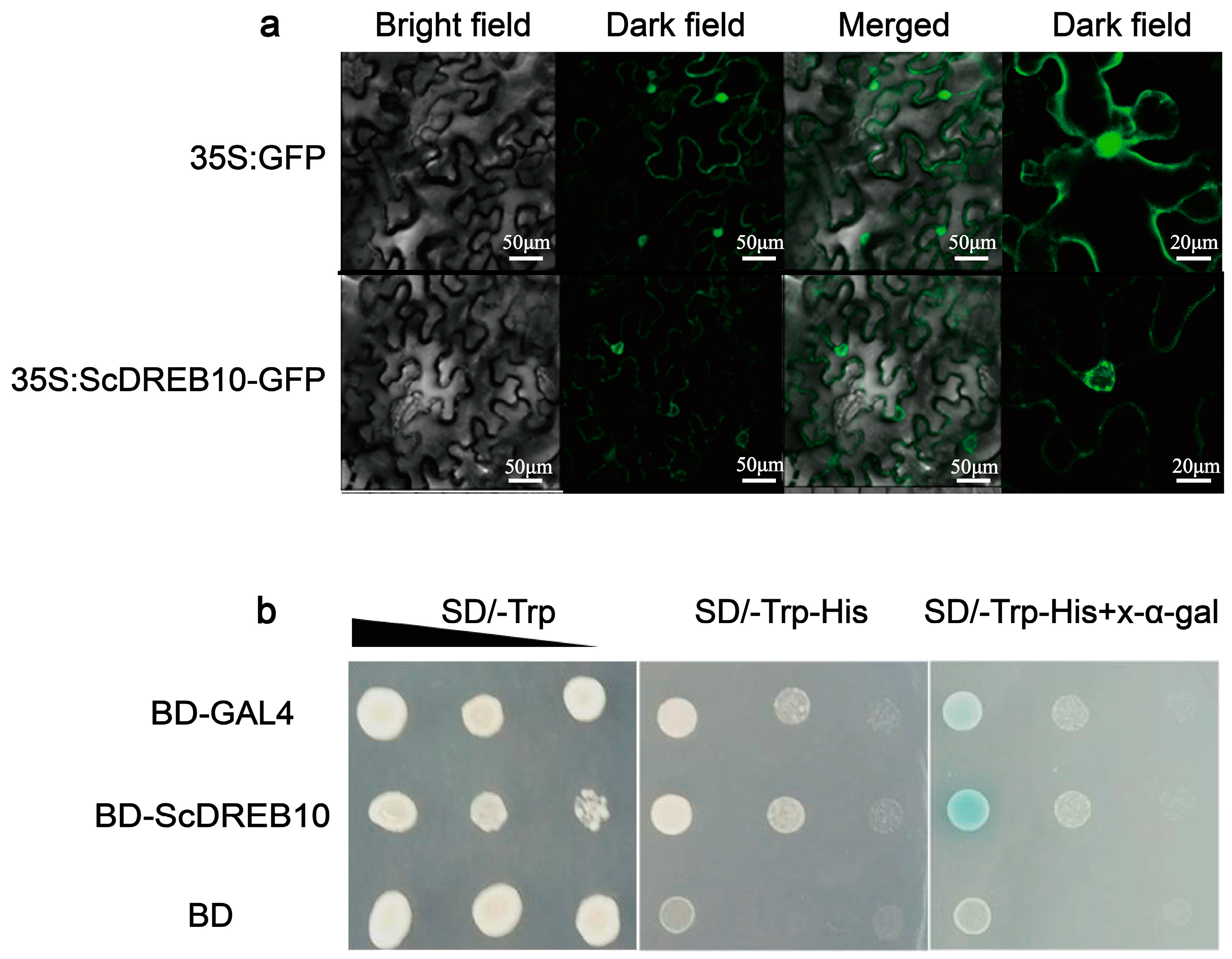
3.2. Localization and Transactivation Activity Analysis of the ScDREB10 Protein
3.3. Overexpression of ScDREB10 Increased the Osmotic and Salt Stress Tolerances in Arabidopsis at the Germination Stage
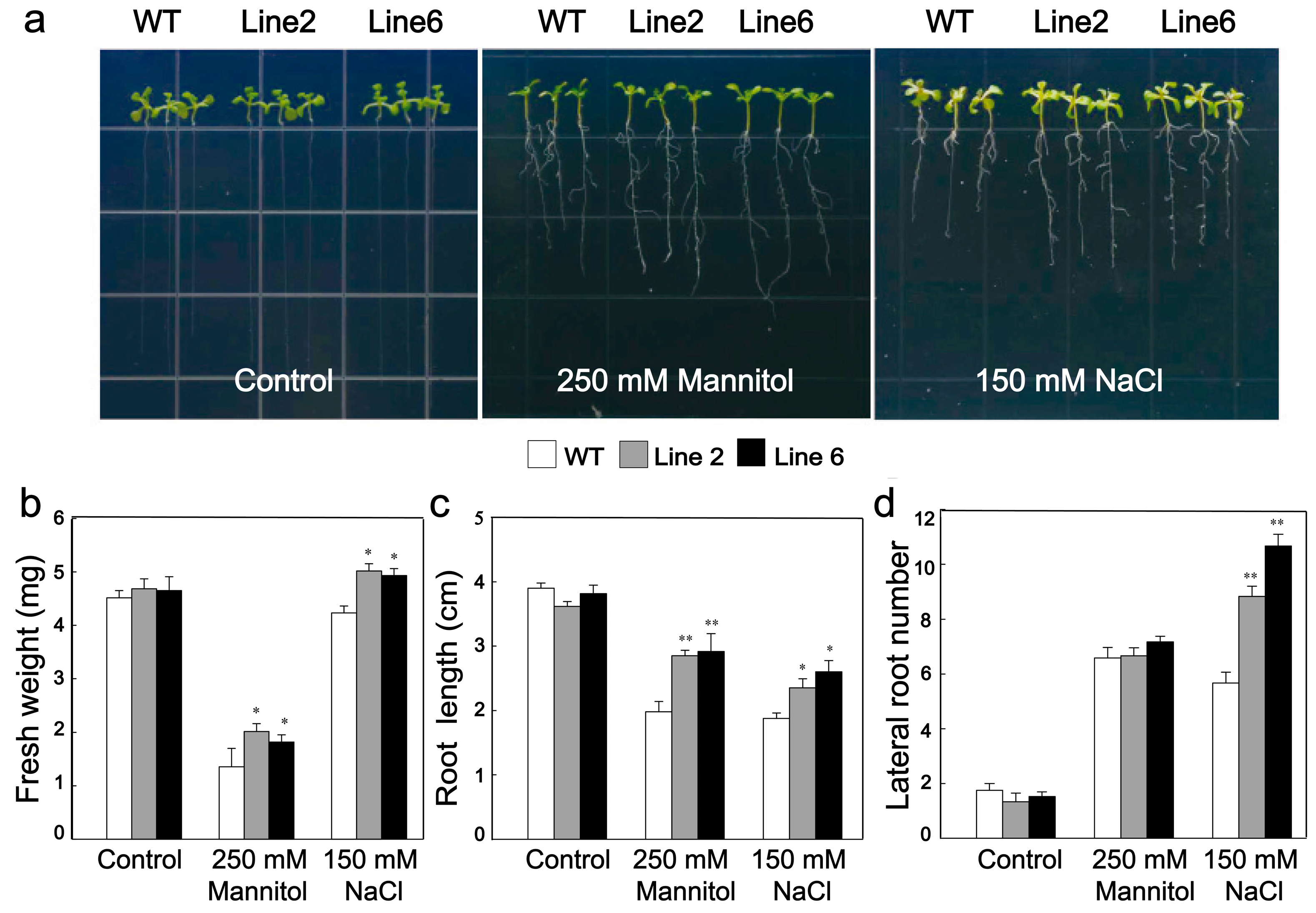
3.4. Overexpression of ScDREB10 Improved the Osmotic and Salt Stress Tolerances of Transgenic Arabidopsis at Seedling Stage
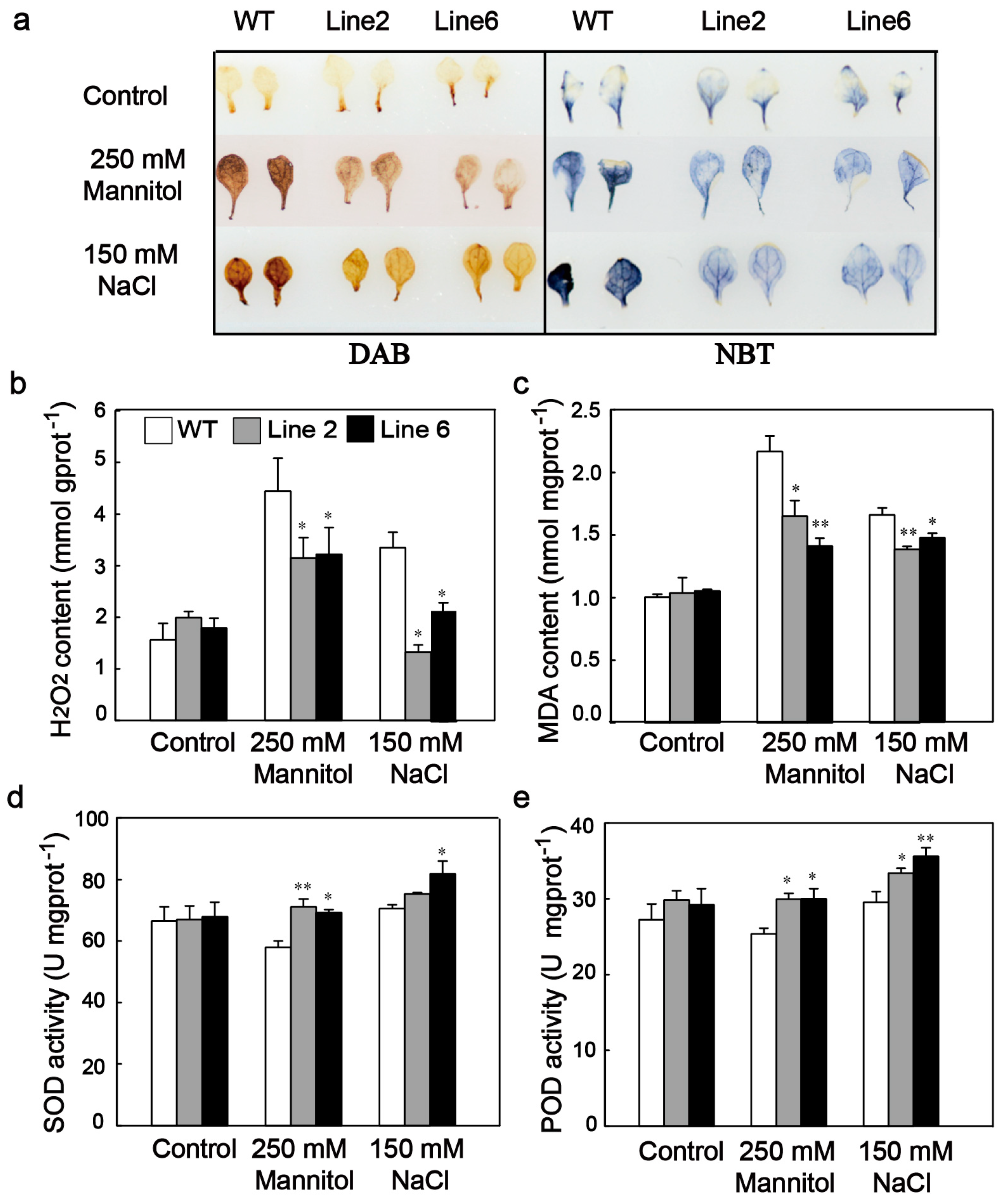
3.5. ScDREB10 Improved ROS Scavenging Capability and Decreased the ROS Damage of Transgenic Arabidopsis under Osmotic and Salt Stresses
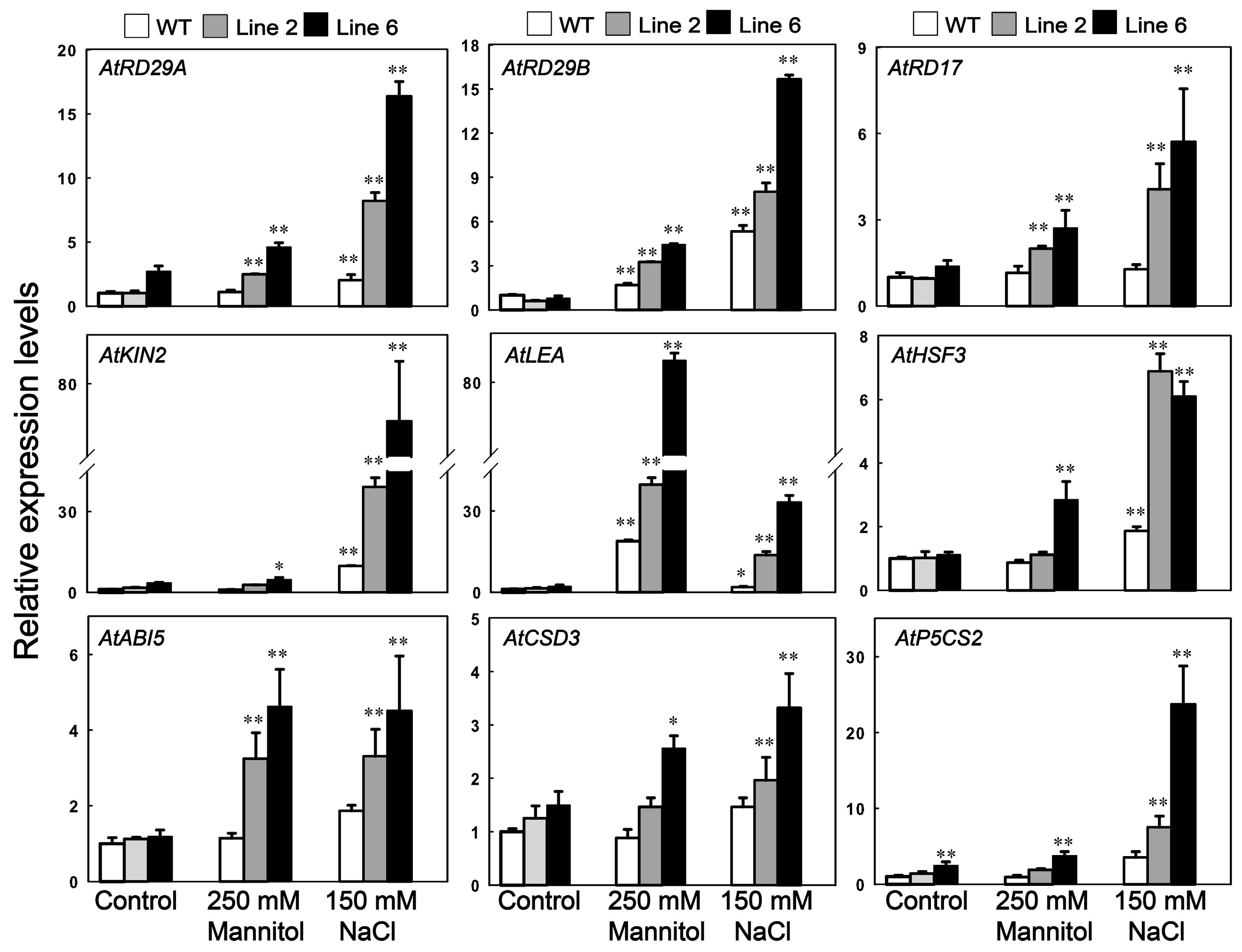
3.6. Analysis of the Potential Downstream Genes Regulated by ScDREB10 in Response to Osmotic and Salt Stresses
4. Discussion
4.1. ScDREB10 is an A-5c Type of DREB Gene Which Can Improve Plant Osmotic and Salt Tolerance without Growth Penalty
4.2. ScDREB10 Accumulation Enhances ROS Scavenging Ability and Upregulates the Expression of Many Stress-Related Genes under Stress Conditions
4.3. ScDREB10 may Exert a Regulatory Function near the Nuclear Membrane
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Silveira, J.A.G.; Carvalho, F.E.L. Proteomics, photosynthesis and salt resistance in crops: An integrative view. J. Proteom. 2016, 143, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Jin, S.X.; Min, L.; He, X.; Li, Y.Y.; Zhu, Y.; Nie, Y.C.; Zhang, X.L.; Zhu, L.F. The Sophora Alopecuroid H+-PPase gene SaVP1 confers multiple abiotic stress tolerance in Arabidopsis. Plant Mol. Biol. Rep. 2015, 33, 923–930. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Valliyodan, B.; Nguyen, H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006, 9, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Jin, L.G.; Liu, J.Y. Identification and characterization of the novel gene GhDBP2 encoding a DRE-binding protein from cotton (Gossypium hirsutum). J. Plant Physiol. 2008, 165, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Matsukura, S.; Mizoi, J.; Yoshida, T.; Todaka, D.; Ito, Y.; Maruyama, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genom. 2010, 283, 185–196. [Google Scholar] [CrossRef]
- Mizoi, J.; Ohori, T.; Moriwaki, T.; Kidokoro, S.; Todaka, D.; Maruyama, K.; Kusakabe, K.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 2013, 161, 346–361. [Google Scholar] [CrossRef]
- Qin, F.; Kakimoto, M.; Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Tran, L.S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007, 50, 54–69. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.M.; Dong, J.; Liu, Y.; Gao, H.W. A Novel Dehydration-Responsive Element-Binding protein from Caragana korshinskii is involved in the response to multiple abiotic stresses and enhances stress tolerance in transgenic tobacco. Plant Mol. Biol. Rep. 2010, 28, 664–675. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.; Li, H.; Wang, Y.; Zhang, Y.; Wood, A.J. EsDREB2B, a novel truncated DREB2-type transcription factor in the desert legume Eremosparton songoricum, enhances tolerance to multiple abiotic stresses in yeast and transgenic tobacco. BMC Plant Biol. 2014, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.Y.; Yu, L.L.; Zhang, K.M.; Zhao, Y.L.; Guo, Y.C.; Gao, C.Q. A ThDREB gene from Tamarix hispida improved the salt and drought tolerance of transgenic tobacco and T. hispida. Plant Physiol. Biochem. 2017, 113, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Gupta, K.; Lopato, S.; Agarwal, P. Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 2017, 68, 2135–2148. [Google Scholar] [CrossRef] [PubMed]
- Lata, C.M. Prasad, Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Wang, M.L.; Lynch, T.J.; Rao, S.; Goodman, H.M. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 1998, 10, 1043–1054. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.; Gao, B.; Liang, Y.; Yang, H.; Wang, Y.; Wood, A.J. Transcriptome-wide identification, classification, and characterization of AP2/ERF family genes in the desert moss Syntrichia caninervis. Front. Plant Sci. 2017, 8, 262. [Google Scholar] [CrossRef]
- Zhang, Y.M. The microstructure and formation of biological soil crusts in their early developmental stage. Chin. Sci. Bull. 2005, 50, 117–121. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, D.; Li, H.; Dong, L.; Lan, H. Ectopic overexpression of the aldehyde dehydrogenase ALDH21 from Syntrichia caninervis in tobacco confers salt and drought stress tolerance. Plant Physiol. Biochem. 2015, 95, 83–91. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.; Li, H.; Gao, B.; Yang, H.; Zhang, Y.; Wood, A.J. Characterization of reference genes for RT-qPCR in the desert moss Syntrichia caninervis in response to abiotic stress and desiccation/rehydration. Front. Plant Sci. 2015, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, D.; Wang, J.; Wood, A.J.; Zhang, Y. Molecular cloning of a stress-responsive aldehyde dehydrogenase gene ScALDH21 from the desiccation-tolerant moss Syntrichia caninervis and its responses to different stresses. Mol. Biol. Rep. 2012, 39, 2645–2652. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Zhang, D.; Li, X.; Yang, H.; Wood, A.J. De novo assembly and characterization of the transcriptome in the desiccation-tolerant moss Syntrichia caninervis. BMC Res. Notes 2014, 7, 490. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, D.; Li, X.; Guan, K.; Yang, H. Novel DREB A-5 subgroup transcription factors from desert moss (Syntrichia caninervis) confers multiple abiotic stress tolerance to yeast. J. Plant Physiol. 2016, 194, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Li, X.; Zhang, D.; Gao, B.; Yang, H.; Wang, Y.; Guan, K.; Wood, A.J. ScDREB8, a novel A-5 type of DREB gene in the desert moss Syntrichia caninervis, confers salt tolerance to Arabidopsis. Plant Physiol. Biochem. 2017, 120, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Bechtold, N.; Pelletier, G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998, 82, 259. [Google Scholar]
- Shi, H.; Wang, Y.; Cheng, Z.; Ye, T.; Chan, Z. Analysis of natural variation in bermudagrass (Cynodon dactylon) reveals physiological responses underlying drought tolerance. PLoS ONE 2012, 7, e53422. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.T.; Ye, T.T.; Chen, F.F.; Cheng, Z.M.; Wang, Y.P.; Yang, P.F.; Zhang, Y.S.; Chan, Z.L. Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: Effect on arginine metabolism and ROS accumulation. J. Exp. Bot. 2013, 64, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.G.; Toyota, M.; Kim, S.H.; Hilleary, R.; Gilroy, S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 6497–6502. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Ji, X.Y.; Nie, X.G.; Qu, M.; Zheng, L.; Tan, Z.L.; Zhao, H.M.; Huo, L.; Liu, S.N.; Zhang, B.; et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 2015, 207, 692–709. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, T.; Maruyama, K.; Obara, M.; Fukutani, A.; Yamaguchi-Shinozaki, K.; Ito, Y.; Kumashiro, T. Expression of Arabidopsis DREB1C improves survival, growth, and yield of upland New Rice for Africa (NERICA) under drought. Mol. Breed. 2013, 31, 255–264. [Google Scholar] [CrossRef]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006, 47, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef]
- Dong, C.J.; Liu, J.Y. The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 2010, 10, 47. [Google Scholar] [CrossRef]
- Zhao, X.J.; Lei, H.J.; Zhao, K.; Yuan, H.Z.; Li, T.H. Isolation and characterization of a dehydration responsive element binding factor MsDREBA5 in Malus sieversii Roem. Sci. Hortic.-Amsterdam 2012, 142, 212–220. [Google Scholar] [CrossRef]
- Bouaziz, D.; Pirrello, J.; Amor, H.B.; Hammami, A.; Charfeddine, M.; Dhieb, A.; Bouzayen, M.; Gargouri-Bouzid, R. Ectopic expression of dehydration responsive element binding proteins (StDREB2) confers higher tolerance to salt stress in potato. Plant Physiol. Biochem. 2012, 60, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, Z.; Xia, L.; Li, L.; Cheng, X.; Dong, J.; Wang, Q.; Ma, Y. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J. Exp. Bot. 2009, 60, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhong, N.Q.; Wang, G.L.; Li, L.J.; Liu, X.L.; He, Y.K.; Xia, G.X. Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens. Planta 2007, 226, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Liu, J.Y. A cotton dehydration responsive element binding protein functions as a transcriptional repressor of DRE-mediated gene expression. Biochem. Biophys. Res. Commun. 2006, 343, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Q.Y.; Cheng, X.G.; Xu, Z.S.; Li, L.C.; Ye, X.G.; Xia, L.Q.; Ma, Y.Z. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007, 353, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.T.; Yin, C.C.; Guo, Q.Q.; Zhou, M.L.; Wang, Z.L.; Wu, Y.M. A novel DREB transcription factor from Halimodendron halodendron leads to enhance drought and salt tolerance in Arabidopsis. Biol. Plant. 2014, 59, 74–82. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Xu, M.Y.; Li, L.H.; Fan, Y.L.; Wan, J.M.; Wang, L. ZmCBF3 overexpression improves tolerance to abiotic stress in transgenic rice (Oryza sativa) without yield penalty. Plant Cell Rep. 2011, 30, 1949–1957. [Google Scholar] [CrossRef]
- Oh, S.J.; Song, S.I.; Kim, Y.S.; Jang, H.J.; Kim, S.Y.; Kim, M.; Kim, Y.K.; Nahm, B.H.; Kim, J.K. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005, 138, 341–351. [Google Scholar] [CrossRef]
- Shukla, R.K.; Raha, S.; Tripathi, V.; Chattopadhyay, D. Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol. 2006, 142, 113–123. [Google Scholar] [CrossRef]
- Shen, Y.G.; Zhang, W.K.; He, S.J.; Zhang, J.S.; Liu, Q.; Chen, S.Y. An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Appl. Genet. 2003, 106, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Baxter, C.J.; Redestig, H.; Schauer, N.; Repsilber, D.; Patil, K.R.; Nielsen, J.; Selbig, J.; Liu, J.; Fernie, A.R.; Sweetlove, L.J. The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol. 2007, 143, 312–325. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [PubMed]
- Dahro, B.; Wang, F.; Peng, T.; Liu, J.H. PtrA/NINV, an alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant Biol. 2016, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.Z.; Wang, Q.B.; Kwon, S.Y.; Kwak, S.S.; Su, W.A. Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. J. Plant Physiol. 2005, 162, 465–472. [Google Scholar] [CrossRef]
- Koussevitzky, S.; Suzuki, N.; Huntington, S.; Armijo, L.; Sha, W.; Cortes, D.; Shulaev, V.; Mittler, R. Ascorbate Peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J. Biol. Chem. 2008, 283, 34197. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Jin, Y.H.; Niu, B.; Feng, K.Y.; Lu, W.C.; Cai, Y.D.; Li, G.Z. Predicting subcellular localization with AdaBoost Learner. Protein Peptide Lett. 2008, 15, 286–289. [Google Scholar]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, L.; Zhang, F.Y.; Jiang, W.M.; Shen, Q.; Zhang, L.D.; Lv, Z.Y.; Wang, G.F.; Tang, K.X. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 2013, 198, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, Y.S.; Kim, S.G.; Jung, J.H.; Woo, J.C.; Park, C.M. Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 2011, 156, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Inzé, A.; Vanderauwera, S.; Hoeberichts, F.A.; Vandorpe, M.; Van, G.T.; Van, B.F. A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 2012, 35, 308–320. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Liang, Y.; Gao, B.; Mijiti, M.; Bozorov, T.A.; Yang, H.; Zhang, D.; Wood, A.J. ScDREB10, an A-5c type of DREB Gene of the Desert Moss Syntrichia caninervis, Confers Osmotic and Salt Tolerances to Arabidopsis. Genes 2019, 10, 146. https://doi.org/10.3390/genes10020146
Li X, Liang Y, Gao B, Mijiti M, Bozorov TA, Yang H, Zhang D, Wood AJ. ScDREB10, an A-5c type of DREB Gene of the Desert Moss Syntrichia caninervis, Confers Osmotic and Salt Tolerances to Arabidopsis. Genes. 2019; 10(2):146. https://doi.org/10.3390/genes10020146
Chicago/Turabian StyleLi, Xiaoshuang, Yuqing Liang, Bei Gao, Meiheriguli Mijiti, Tohir A. Bozorov, Honglan Yang, Daoyuan Zhang, and Andrew J. Wood. 2019. "ScDREB10, an A-5c type of DREB Gene of the Desert Moss Syntrichia caninervis, Confers Osmotic and Salt Tolerances to Arabidopsis" Genes 10, no. 2: 146. https://doi.org/10.3390/genes10020146
APA StyleLi, X., Liang, Y., Gao, B., Mijiti, M., Bozorov, T. A., Yang, H., Zhang, D., & Wood, A. J. (2019). ScDREB10, an A-5c type of DREB Gene of the Desert Moss Syntrichia caninervis, Confers Osmotic and Salt Tolerances to Arabidopsis. Genes, 10(2), 146. https://doi.org/10.3390/genes10020146







