Morphological Characterization of Flower Buds Development and Related Gene Expression Profiling at Bud Break Stage in Heterodichogamous Cyclocarya paliurus (Batal.) lljinskaja
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant for Sample Collection
2.2. RNA Extraction, cDNA Synthesis, and Sequencing
2.3. Data Filtering, De Novo Assembly, and Functional Annotation
2.4. Differentially Expressed Genes Between Female and Male Buds in Two Mating Types
2.5. Quantitative Real-Time PCR Verification
3. Results
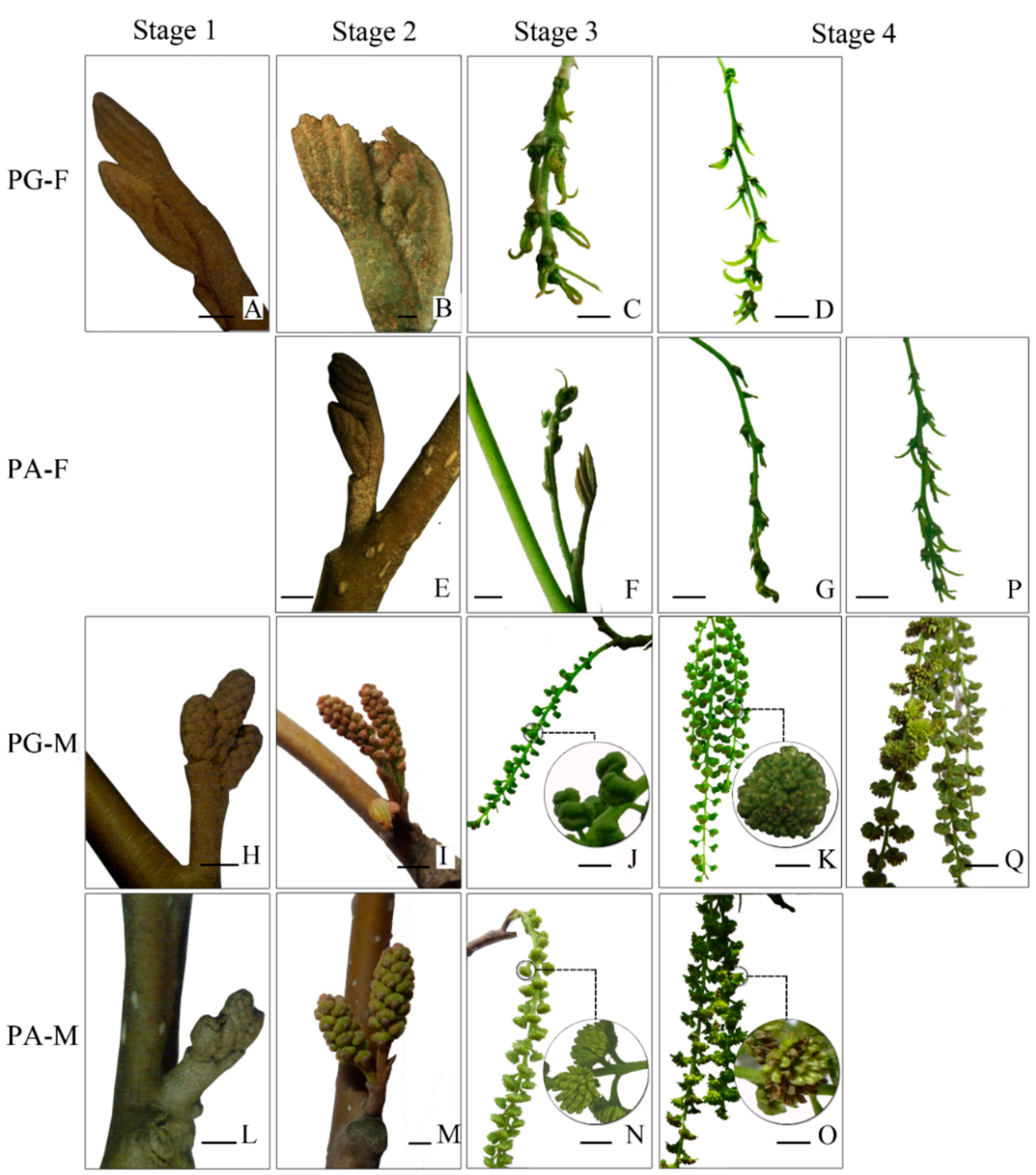
3.1. Morphological Characteristics of Floral Bud Development
3.2. Sequencing, Assembly, and Annotation
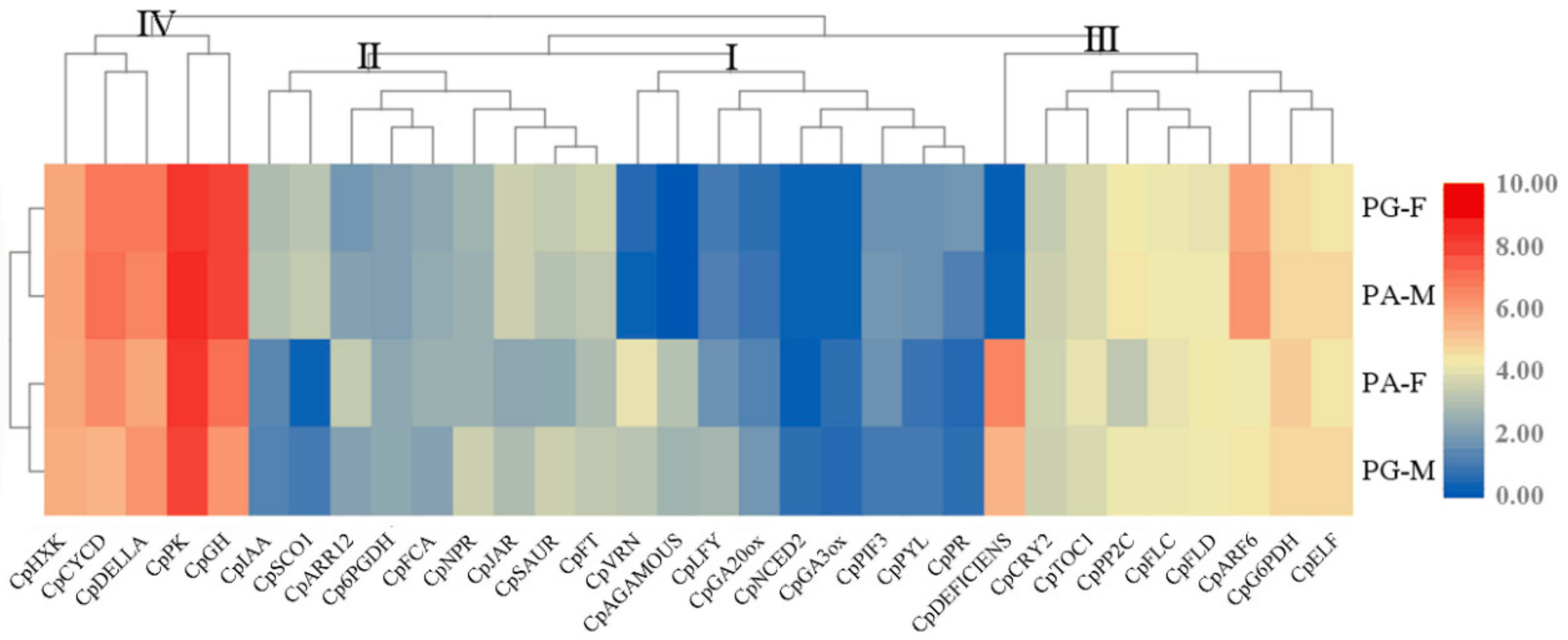
3.3. Expression of Major Genes Related to Flower Differentiation
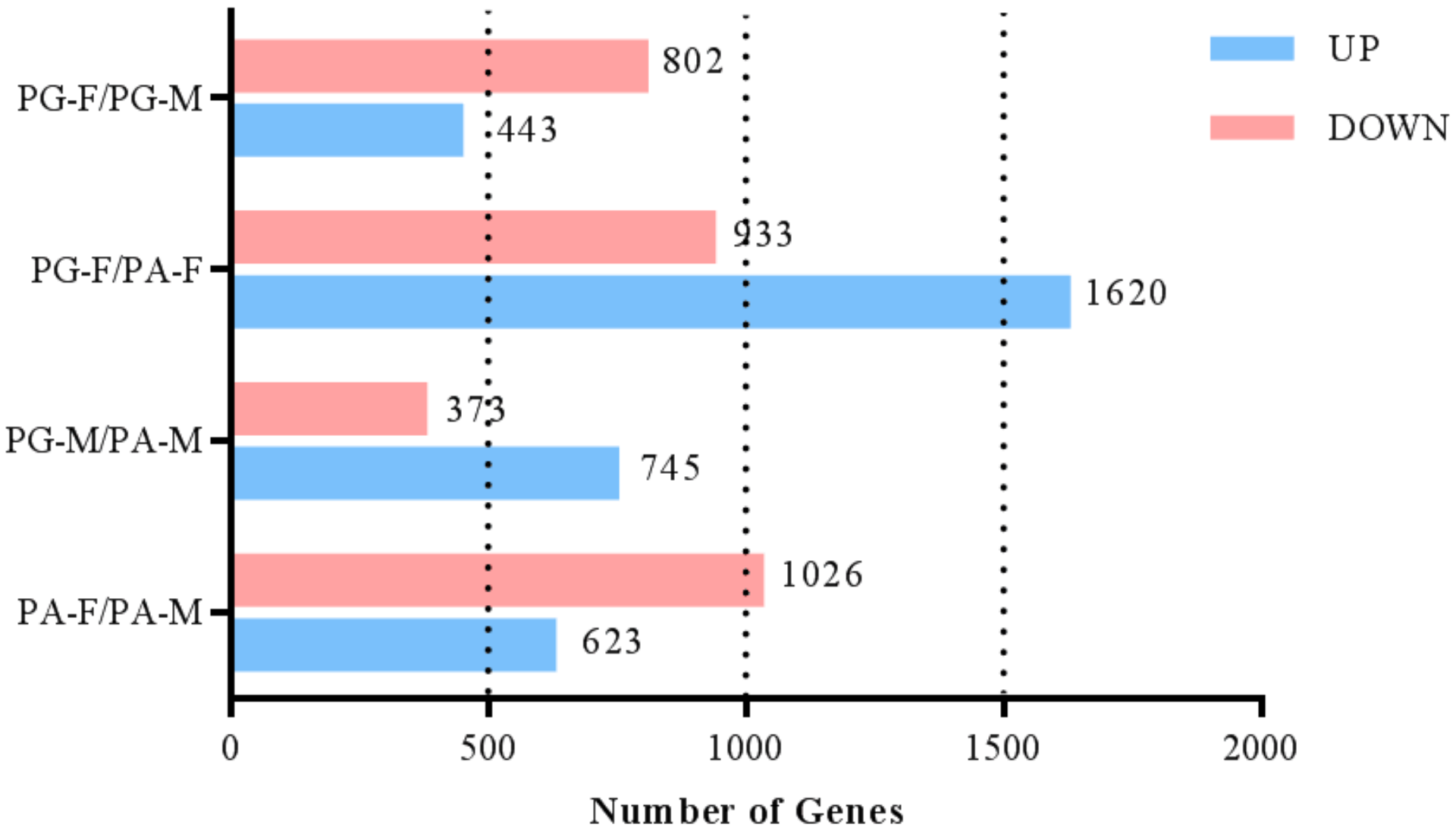
3.4. Identification of Differentially Expressed Genes
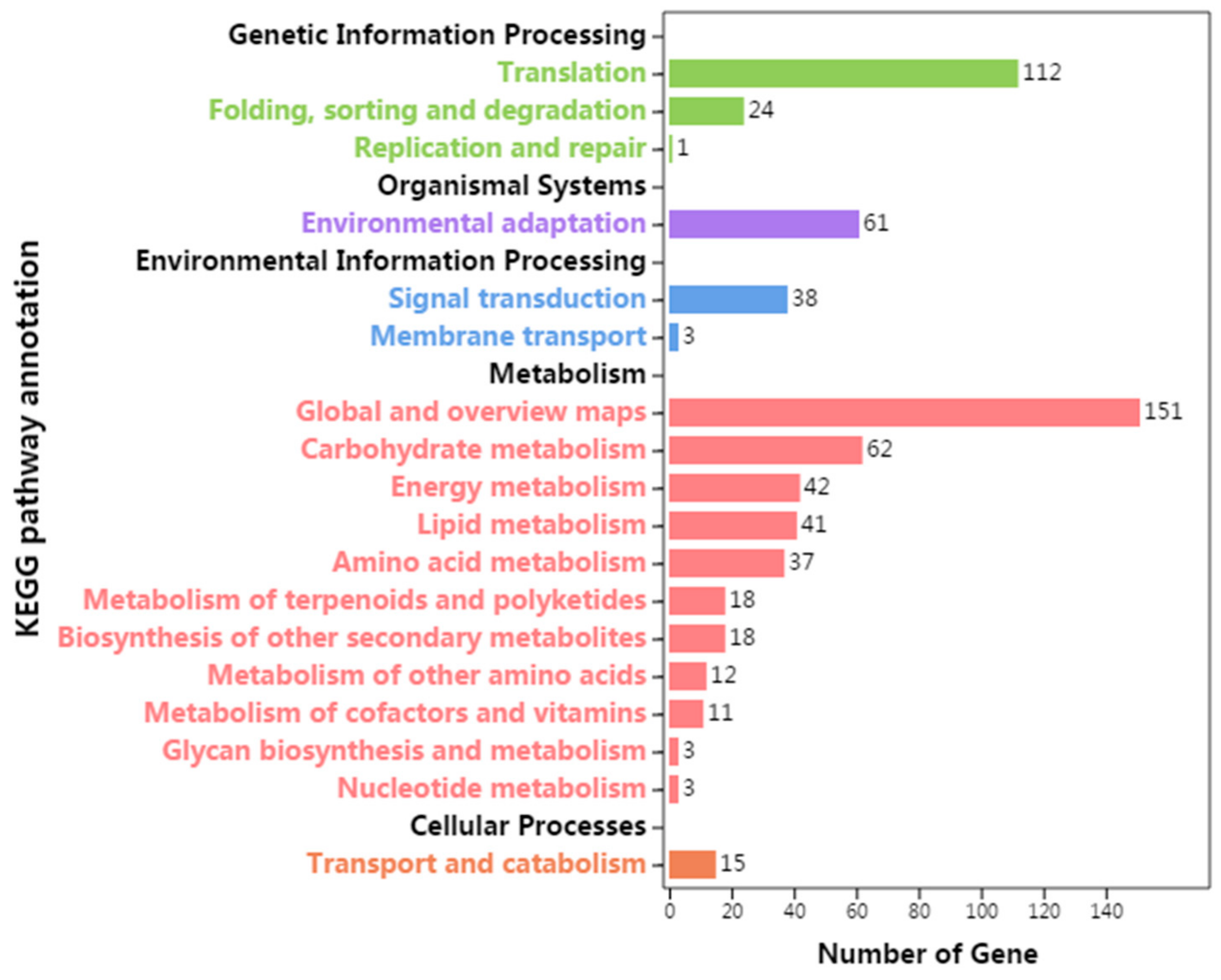
3.5. KEGG Pathway Annotation of Differentially Expressed Genes
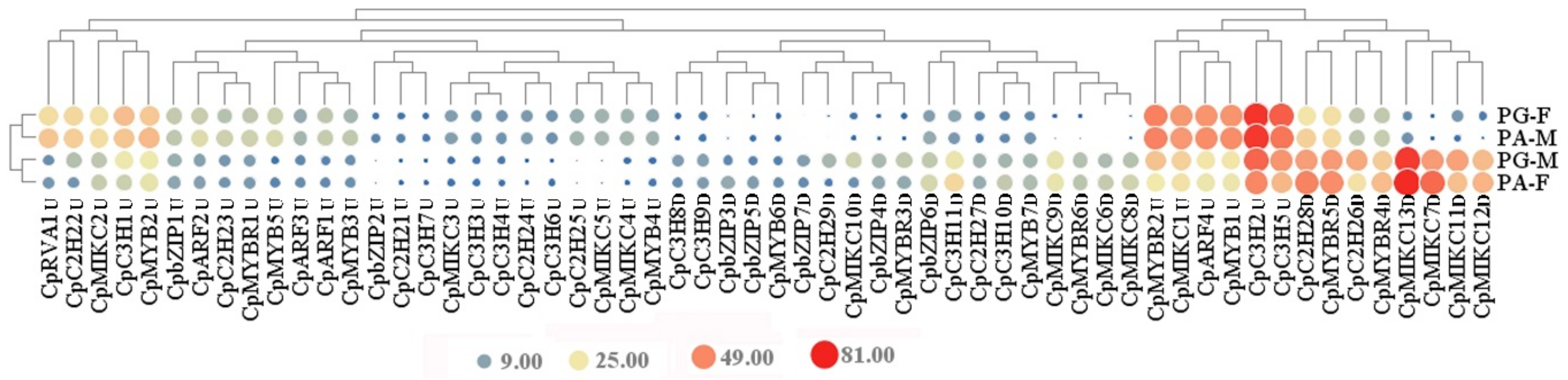
3.6. Major Differentially Expressed Transcription Factors
3.7. Validation of RNA-Seq Result by qRT-PCR
4. Discussion
4.1. Expression Pattern of Major Genes in Different Pathways of Forming Flowers
4.2. Expression Pattern of TFs Related to Flowering from DEGs
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kikuchi, S.; Shibata, M.; Tanaka, H.; Yoshimaru, H.; Niiyama, K. Analysis of the disassortative mating pattern in a heterodichogamous plant, Acer mono Maxim. using microsatellite markers. Plant Ecol. 2009, 204, 43–54. [Google Scholar] [CrossRef]
- Liu, J.J.; Mao, X.; Li, X.C. Molecular mechanism of controlling flower formation by photoperiod inducement in plants. J. Nanjing For. Univ. 2016, 40, 147–154. [Google Scholar]
- Fukuhara, T.; Tokumaru, S.I. Inflorescence dimorphism, heterodichogamy and thrips pollination in Platycarya strobilacea (Juglandaceae). Ann. Bot. 2014, 113, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.P.; Shangguan, X.C.; Chen, J.G.; Zhao, Q.; Li, D.M. Growth and triterpenic acid accumulation of Cyclocarya paliurus cell suspension cultures. Biotechnol. Bioprocess Eng. 2013, 18, 606–614. [Google Scholar] [CrossRef]
- Yang, H.M.; Yin, Z.Q.; Zhao, M.G.; Jiang, C.H.; Zhang, J.; Pan, K. Pentacyclic triterpenoids from Cyclocarya paliurus and their antioxidant activities in FFA-induced HepG2 steatosis cells. Phytochemistry 2018, 151, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.W.; Zhai, L.X.; Huang, T.; Peng, J.; Hu, D.; Xiao, H.T.; Wen, B.; Lin, C.Y.; Zhao, L.; Bian, Z.X. Identification of α-glucosidase inhibitors from: Cyclocarya paliurus tea leaves using UF-UPLC-Q/TOF-MS/MS and molecular docking. Food Funct. 2019, 10, 1893–1902. [Google Scholar] [CrossRef]
- Xie, J.H.; Wang, W.J.; Dong, C.J.; Huang, L.X.; Wang, H.; Li, C.; Nie, S.; Xie, M.Y. Protective effect of flavonoids from Cyclocarya paliurus leaves against carbon tetrachloride-induced acute liver injury in mice. Food Chem. Toxicol. 2018, 119, 392–399. [Google Scholar] [CrossRef]
- Li, X.C.; Fu, X.X.; Shang, X.L.; Yang, W.X.; Fang, S.Z. Natural population structure and genetic differentiation for heterodicogamous plant: Cyclocarya paliurus (Batal.) Iljinskaja (Juglandaceae). Tree Genet. Genomes 2017, 13. [Google Scholar] [CrossRef]
- Mao, X.; Liu, J.J.; Li, X.C.; Qin, J.; Fu, X.X. Flowering biological characteristics and mating system in immature plantations of heterodichogamous Cyclocarya paliurus. J. Nanjing For. Univ. 2016, 40, 47–55. [Google Scholar]
- Mao, X.; Fu, X.X.; Huang, P.; Chen, X.L.; Qu, Y.Q. Heterodichogamy, pollen viability, and seed set in a population of polyploidy Cyclocarya paliurus (Batal) Iljinskaja (Juglandaceae). Forests 2019, 10, 347. [Google Scholar] [CrossRef]
- Fu, X.X.; Feng, L.; Fang, S.Z.; Mao, J. Observation on flowering habits and anatomy of stamen development in Cyclocarya paliurus. J. Nanjing For. Univ. 2010, 34, 67–71. [Google Scholar]
- Fu, X.X.; Feng, L.; Shang, X.L.; Yang, W.X.; Fang, S.Z. Observation of morphological and anatomical characters on staminate and pistillate flower differentiation in Cyclocarya paliurus. J. Nanjing For. Univ. 2011, 35, 17–22. [Google Scholar]
- Sun, Y.Y.; Wang, G.D.; Li, Y.X.; Jiang, L.; Yang, Y.X.; Guan, S.X. De novo transcriptome sequencing and comparative analysis to discover genes related to floral development in Cymbidium faberi Rolfe. SpringerPlus 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; De Montaigu, A.; Coupland, G. SnapShot: Control of Flowering in Arabidopsis. Cell 2010, 141. [Google Scholar] [CrossRef]
- King, R.W.; Moritz, T.; Evans, L.T.; Martin, J.; Andersen, C.H.; Blundell, C.; Kardailsky, I.; Chandler, P.M. Regulation of flowering in the long-day grass Lolium temulentum by gibberellins and the FLOWERING LOCUS T gene. Plant Physiol. 2006, 141, 498–507. [Google Scholar] [CrossRef]
- Wang, H.Z.; Hu, B.; Chen, G.P.; Shi, N.N.; Zhao, Y.; Yin, Q.C.; Liu, J.J. Application of Arabidopsis AGAMOUS second intron for the engineered ablation of flower development in transgenic tobacco. Plant Cell Rep. 2008, 27, 251–259. [Google Scholar] [CrossRef]
- Zhang, M.Z.; Ye, D.; Wang, L.L.; Pang, J.L.; Zhang, Y.H.; Zheng, K.; Bian, H.W.; Han, N.; Pan, J.W.; Wang, J.H.; et al. Overexpression of the cucumber LEAFY homolog CFL and hormone treatments alter flower development in gloxinia (Sinningia speciosa). Plant Mol. Biol. 2008, 67, 419–427. [Google Scholar] [CrossRef]
- Marsch-Martínez, N.; De Folter, S. Hormonal control of the development of the gynoecium. Curr. Opin. Plant Biol. 2016, 29, 104–114. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. The ABC model of flower development: Then and now. Development 2012, 139, 4095–4098. [Google Scholar] [CrossRef]
- Gomez, J.F.; Talle, B.; Wilson, Z.A. Anther and pollen development: A conserved developmental pathway. J. Integr. Plant Biol. 2015, 57, 876–891. [Google Scholar] [CrossRef] [Green Version]
- Pajoro, A.; Biewers, S.; Dougali, E.; Valentim, F.L.; Mendes, M.A.; Porri, A.; Coupland, G.; Van De Peer, Y.; Van Dijk, A.D.J.; Colombo, L.; et al. The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: A two decades history. J. Exp. Bot. 2014, 65, 4731–4745. [Google Scholar] [CrossRef] [PubMed]
- Vito, S.P.; Naiyan, L. Pistillate flower differentiation in English walnut (Juglans regia L.): A development basis for heterodichogamy. Scientia Horticulturae 1985, 26, 333–338. [Google Scholar]
- Bai, W.N.; Zeng, Y.F.; Zhang, D.Y. Mating patterns and pollen dispersal in a heterodichogamous tree, Juglans mandshurica (Juglandaceae). New Phytol. 2007, 176, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Polito, V.S.; Pinney, K. The relationship between phenology of pistillate flower organogenesis and mode of heterodichogamy in Juglans regia L. (Juglandaceae). Sex. Plant Reprod. 1997, 10, 36–39. [Google Scholar] [CrossRef]
- Huang, P.; Mao, X.; Han, G.; Chen, X.L.; Fu, X.X. Dynamic changes of nutrients during flora development in heterodichogamous Cyclocarya paliurus. J. Nanjing For. Univ. 2018, 42, 1–9. [Google Scholar]
- Zhang, X.-X.; Jiang, C.-H.; Liu, Y.; Lou, D.-X.; Huang, Y.-P.; Gao, M.; Zhang, J.; Yin, Z.-Q.; Pan, K. Cyclocarya paliurus triterpenic acids fraction attenuates kidney injury via AMPK-mTOR-regulated autophagy pathway in diabetic rats. Phytomedicine 2019, 64, 153060. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, J.; Wang, J.; Li, J.; Ouyang, K.; Wang, W. Effects of Cyclocarya paliurus polysaccharide on lipid metabolism-related genes DNA methylation in rats. Int. J. Biol. Macromol. 2019, 123, 343–349. [Google Scholar] [CrossRef]
- Xuan, T.Y.; Tan, J.; Sun, H.H.; Yang, C.; Lv, W.Y.; Zhang, J.H.; Zhang, K.Q.; Nie, Z.Q.; Ye, Z.J.; He, X.A.; et al. Cyclocarioside O-Q, three novel seco-dammarane triterpenoid glycosides from the leaves of Cyclocarya paliurus. Nat. Prod. Res. 2019, 14, 1–7. [Google Scholar] [CrossRef]
- Zhou, M.; Lin, Y.; Fang, S.; Liu, Y.; Shang, X. Phytochemical content and antioxidant activity in aqueous extracts of Cyclocarya paliurus leaves collected from different populations. PeerJ 2019, 2019, 1–18. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y. A study on the developmental characteristics of walnut buds. Acta Hortic. Sin. 1965, 4, 61–69. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fang, S.; Yang, W.; Shang, X.; Fu, X. Light quality affects flavonoid production and related gene expression in Cyclocarya paliurus. J. Photochem. Photobiol. B Biol. 2018, 179, 66–73. [Google Scholar] [CrossRef] [PubMed]
- He, H.H.; Liang, G.P.; Lu, S.X.; Wang, P.P.; Liu, T.; Ma, Z.H.; Zuo, C.W.; Sun, X.M.; Chen, B.H.; Mao, J. Genome-Wide Identification and Expression Analysis of GA2ox, GA3ox, and GA20ox Are Related to Gibberellin Oxidase Genes in Grape (Vitis vinifera L.). Genes 2019, 10, 680. [Google Scholar] [CrossRef]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019, 287. [Google Scholar] [CrossRef]
- Pidal, B.; Yan, L.; Fu, D.; Zhang, F.; Tranquilli, G.; Dubcovsky, J. The CArG-box located upstream from the transcriptional start of wheat vernalization gene VRN1 is not necessary for the vernalization response. J. Hered. 2009, 100, 355–364. [Google Scholar] [CrossRef]
- Yu, D.; Qanmber, G.; Lu, L.; Wang, L.; Li, J.; Yang, Z.; Liu, Z.; Li, Y.; Chen, Q.; Mendu, V.; et al. Genome-wide analysis of cotton GH3 subfamily II reveals functional divergence in fiber development, hormone response and plant architecture. BMC Plant Biol. 2018, 18. [Google Scholar] [CrossRef]
- Cortleven, A.; Schmülling, T. Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 2015, 66, 4999–5013. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.J.; Fu, T.Y.; Yang, S.L.; Hsieh, H.L. FIN219/JAR1 and cryptochrome1 antagonize each other to modulate photomorphogenesis under blue light in Arabidopsis. PLoS Genet. 2018, 14, e1007248. [Google Scholar] [CrossRef]
- Pokotylo, I.; Kravets, V.; Ruelland, E. Salicylic Acid Binding Proteins (SABPs): The Hidden Forefront of Salicylic Acid Signalling. Int. J. Mol. Sci. 2019, 20, 4377. [Google Scholar] [CrossRef] [PubMed]
- Hung, F.Y.; Chen, F.F.; Li, C.; Chen, C.; Chen, J.H.; Cui, Y.; Wu, K. The LDL1/2-HDA6 histone modification complex interacts with TOC1 and regulates the core circadian clock components in Arabidopsis. Front. Plant Sci. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Golicz, A.A. Analysis of the quinoa genome reveals conservation and divergence of the flowering pathways. Funct. Integr. Genom. 2019, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Granot, D.; Kelly, G.; Stein, O.; David-Schwartz, R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 2014, 65, 809–819. [Google Scholar] [CrossRef]
- Sung, S.S.; Xu, D.P.; Galloway, C.M.; Black, C.C. A reassessment of glycolysis and gluconeogenesis in higher plants. Physiol. Plant. 1988, 72. [Google Scholar] [CrossRef]
- Arro, J.; Yang, Y.; Song, G.Q.; Zhong, G.Y. RNA-Seq reveals new della targets and regulation in transgenic GA-insensitive grapevines. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar] [CrossRef]
- Garza-Aguilar, S.M.; Lara-Núñez, A.; García-Ramírez, E.; Vázquez-Ramos, J.M. Modulation of CycD3;1-CDK complexes by phytohormones and sucrose during maize germination. Physiol. Plant 2017, 160, 84–97. [Google Scholar] [CrossRef]
- Manchado-Rojo, M.; Delgado-Benarroch, L.; Roca, M.J.; Weiss, J.; Egea-Cortines, M. Quantitative levels of Deficiens and Globosa during late petal development show a complex transcriptional network topology of B function. Plant J. 2012, 72, 294–307. [Google Scholar] [CrossRef]
- Wen, J.; Guo, P.; Ke, Y.; Liu, M.; Li, P.; Wu, Y.; Ran, F.; Wang, M.; Li, J.; Du, H. The auxin response factor gene family in allopolyploid Brassica napus. PLoS ONE 2019, 14, e0214885. [Google Scholar] [CrossRef]
- Kruger, N.J.; Schaewen, A. Von The oxidative pentose phosphate pathway: Structure and organization. Curr. Opin. Plant Biol. 2003, 6, 236–246. [Google Scholar] [CrossRef]
- Singh, S.; Srivastava, P.K. Purification and Characterization of Glucose-6-Phosphate Dehydrogenase from Pigeon Pea (Cajanus cajan) Seeds. Adv. Enzym. Res. 2014, 2, 134–149. [Google Scholar] [CrossRef]
- Dong, J.; Sun, N.; Yang, J.; Deng, Z.; Lan, J.; Qin, G.; He, H.; Deng, X.W.; Irish, V.F.; Chen, H.; et al. The transcription factors tcp4 and pif3 antagonistically regulate organ-specific light induction of saur genes to modulate cotyledon opening during de-etiolation in Arabidopsis. Plant Cell 2019, 31, 1155–1170. [Google Scholar] [CrossRef] [PubMed]
- Field, D.L.; Barrett, S.C.H. Disassortative mating and the maintenance of sexual polymorphism in painted maple. Mol. Ecol. 2012, 21. [Google Scholar] [CrossRef]
- Barrett, S.C.H. The evolution of plant sexual diversity. Nat. Rev. Genet. 2002, 3, 274–284. [Google Scholar] [CrossRef]
- Gleiser, G.; Verdú, M.; Segarra-Moragues, J.G.; González-Martínez, S.C.; Pannell, J.R. Disassortative mating, sexual specialization, and the evolution of gender dimorphism in heterodichogamous Acer opalus. Evolution 2008, 62, 1676–1688. [Google Scholar] [CrossRef]
- Jung, J.H.; Seo, P.J.; Ahn, J.H.; Park, C.M. Arabidopsis RNA-binding Protein FCA regulates microRNA172 processing in thermosensory flowering. J. Biol. Chem. 2012, 287, 16007–16016. [Google Scholar] [CrossRef]
- Angeles-Núñez, J.G.; Tiessen, A. Arabidopsis sucrose synthase 2 and 3 modulate metabolic homeostasis and direct carbon towards starch synthesis in developing seeds. Planta 2010, 232, 701–718. [Google Scholar] [CrossRef]
- Han, G. Studies of hormonal regulation on flower bud differentiation and development in heterodichogamous Cyclocarya paliurus. Nanjing For. Univ. 2018. (In Chinese) [Google Scholar]
- Baek, W.; Lim, C.W.; Luan, S.; Lee, S.C. The RING finger E3 ligases PIR1 and PIR2 mediate PP2CA degradation to enhance abscisic acid response in Arabidopsis. Plant J. 2019. [Google Scholar] [CrossRef]
- Martinez-Zapater, J.M.; Coupland, G.; Dean, C.; Koornneef, M. The Transition to Flowering in Arabidopsis; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1994; pp. 403–433. [Google Scholar]
- Achard, P.; Cheng, H.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; Van Der Straeten, D.; Peng, J.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef]
- Schrader, J.; Moyle, R.; Bhalerao, R.; Hertzberg, M.; Lundeberg, J.; Nilsson, P.; Bhalerao, R.P. Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J. 2004, 40, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Looney, N.E. Hormones and horticulture. Hortscience 1997, 32, 1014–1018. [Google Scholar] [CrossRef]
- Saure, M.C. Dormancy release in deciduous fruit trees. Hortic. Rev. 2011, 7, 239–299. [Google Scholar]
- Zhuang, W.B.; Gao, Z.H.; Wang, L.J.; Zhong, W.J.; Ni, Z.J.; Zhang, Z. Comparative proteomic and transcriptomic approaches to address the active role of GA4 in Japanese apricot flower bud dormancy release. J. Exp. Bot. 2013, 64, 4953–4966. [Google Scholar] [CrossRef]
- Binenbaum, J.; Weinstain, R.; Shani, E. Gibberellin localization and transport in plants. Trends Plant Sci. 2018, 23, 410–421. [Google Scholar] [CrossRef]
- Vachon, G.; Engelhorn, J.; Carles, C.C. Interactions between transcription factors and chromatin regulators in the control of flower development. J. Exp. Bot. 2018, 69, 2461–2471. [Google Scholar] [CrossRef]
- Liu, C.; Xi, W.Y.; Shen, L.S.; Tan, C.P.; Yu, H. Regulation of floral patterning by flowering time genes. Dev. Cell 2009, 16, 711–722. [Google Scholar] [CrossRef]
- Liu, J.; Fu, X.; Dong, Y.; Lu, J.; Ren, M.; Zhou, N.; Wang, C. MIKC C-type MADS-box genes in Rosa chinensis: The remarkable expansion of ABCDE model genes and their roles in floral organogenesis. Hortic. Res. 2018, 5, 25. [Google Scholar] [CrossRef]
- Tian, S.B.; Guo, C.X.; Zheng, C.S. Molecular mechanism of controlling flower formation by photoperiod inducement in plants. Acta Hortic. Sin. 2010, 37, 325–330. [Google Scholar] [CrossRef]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 312, 1600. [Google Scholar] [CrossRef]
- Takatsuji, H.; Mori, M.; Benfey, P.N.; Ren, L.; Chua, N.H. Characterization of a zinc finger DNA-binding protein expressed specifically in Petunia petals and seedlings. EMBO J. 1992, 11, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Takatsuji, H.; Nakamura, N.; Katsumoto, Y. A new family of zinc finger proteins in petunia: Structure, DNA sequence recognition, and floral organ-specific expression. Plant Cell 1994, 6, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Immink, R.G.H.; Kaufmann, K.; Angenent, G.C. The “ABC” of MADS domain protein behaviour and interactions. Semin. Cell Dev. Biol. 2010, 21, 87–93. [Google Scholar] [CrossRef]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Wang, H.; Caruso, L.V.; Downie, A.B.; Perry, S.E. The embryo MADS domain protein Agamous-like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 2004, 16, 1206–1219. [Google Scholar] [CrossRef]
- Zhou, K.Y.; Xu, S.Y.; Wu, Q.T.; Shi, Y.; Wu, Q.J. The research progress of AGAMOUS-like orthologous genes in woody plants. For. Sci. Technol. 2018, 43, 26–30. [Google Scholar]
- Yamane, H.; Ooka, T.; Jotatsu, H.; Hosaka, Y.; Sasaki, R.; Tao, R. Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J. Exp. Bot. 2011, 62, 3481–3488. [Google Scholar] [CrossRef]
- Liu, S.; Xia, X.Y.; Li, B. Cloning and expression analysis of MADS-box gene VcDAM1 related to blueberry flower bud dormancy. Plant Physiol. J. 2017, 53, 1728–1734. [Google Scholar]
- Kitamura, Y.; Takeuchi, T.; Yamane, H.; Tao, R. Simultaneous down-regulation of DORMANCY-ASSOCIATED MADS-box6 and SOC1 during dormancy release in Japanese apricot (Prunus mume) flower buds. J. Hortic. Sci. Biotechnol. 2016, 91, 1–7. [Google Scholar] [CrossRef]
- Lin, M.M.; Chen, J.Y.; Gu, H.; Qi, X.J.; Zhang, W.Y.; Zhang, Y.; Fang, J.B. Isolation of a sweet cherry AGAMOUS-like gene and its expression under gibberellin treatment. Mol. Plant Breed 2014, 12, 1181–1186. [Google Scholar]




| Database | Count | Percentage (%) |
|---|---|---|
| BLASTP | 22,563 | 35.51 |
| BLASTX | 26,158 | 41.17 |
| GO | 25,167 | 39.61 |
| KO | 11,027 | 17.36 |
| NR | 37,375 | 58.83 |
| NT | 21,777 | 34.28 |
| PFAM | 23,464 | 36.93 |
| Prot | 31,330 | 49.31 |
| SignalP | 2154 | 3.39 |
| COG | 11,394 | 17.93 |
| TmHMM | 6488 | 10.21 |
| eggNOG | 13,476 | 21.21 |
| Total annotation | 46,009 | 72.42 |
| Total unigenes | 63,533 | 100 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Mao, X.; Huang, P.; Fang, S. Morphological Characterization of Flower Buds Development and Related Gene Expression Profiling at Bud Break Stage in Heterodichogamous Cyclocarya paliurus (Batal.) lljinskaja. Genes 2019, 10, 818. https://doi.org/10.3390/genes10100818
Chen X, Mao X, Huang P, Fang S. Morphological Characterization of Flower Buds Development and Related Gene Expression Profiling at Bud Break Stage in Heterodichogamous Cyclocarya paliurus (Batal.) lljinskaja. Genes. 2019; 10(10):818. https://doi.org/10.3390/genes10100818
Chicago/Turabian StyleChen, Xiaoling, Xia Mao, Peng Huang, and Shengzuo Fang. 2019. "Morphological Characterization of Flower Buds Development and Related Gene Expression Profiling at Bud Break Stage in Heterodichogamous Cyclocarya paliurus (Batal.) lljinskaja" Genes 10, no. 10: 818. https://doi.org/10.3390/genes10100818




