Scavenger Receptor Structure and Function in Health and Disease
Abstract
:1. Introduction
2. Class A
2.1. Genetics, Protein Structure and Expression

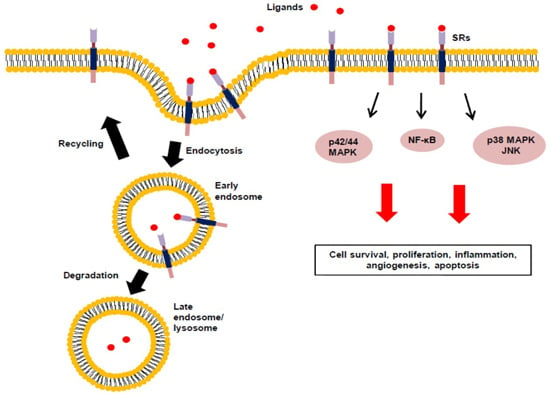
2.2. Signal Transduction, Trafficking and Cell Function
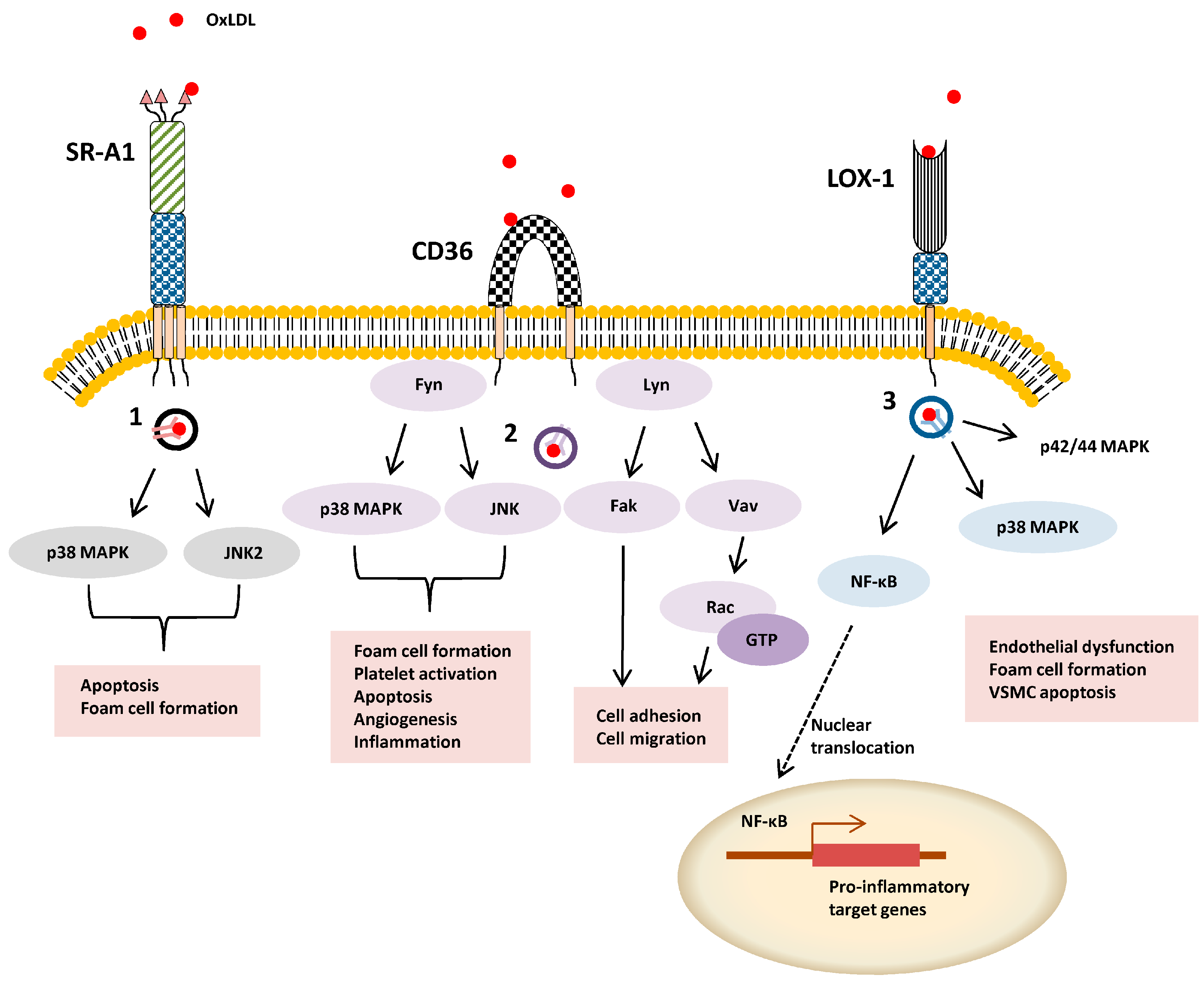
3. Class B
3.1. Genetics, Protein Structure and Expression
3.2. Signal Transduction, Trafficking and Cell Function
4. Class C
5. Class D
6. Class E
6.1. Genetics, Protein Structure and Expression
6.2. Signal Transduction, Trafficking and Cell Function
7. Class F
8.Class G
9. Class H
10. Class I
10.1. Genetics, Protein Structure and Expression
10.2. Signal Transduction, Trafficking and Cell Function
11. Class J
11.1. Genetics, Protein Structure and Expression
11.2. Signal Transduction, Trafficking and Cell Function
12. Biological Roles of SR
12.1. SR and ROS Production
12.2. SR and Apoptosis
12.3. SR and Angiogenesis
13. Concluding Remarks
Acknowledgments
Author contributions
Abbreviations
| SR | scavenger receptor |
| OxLDL | oxidized low-density lipoprotein |
| HDL | high-density lipoprotein |
| CDE | clathrin-dependent endocytosis |
| CIE | clathrin-independent endocytosis |
| AcLDL | acetylated low-density lipoprotein |
| MAPK | mitogen-activated protein kinase |
| JNK | c-Jun N-terminal kinase |
| FAK | focal adhesion kinase |
| AGE | advanced glycation end-products |
| PAMP | pathogen-associated molecular pattern |
| ROS | reactive oxygen species |
| VSMC | vascular smooth muscle cell |
| TLR | Toll-like receptor |
Conflicts of Interest
References
- Prabhudas, M.; Bowdish, D.; Drickamer, K.; Febbraio, M.; Herz, J.; Kobzik, L.; Krieger, M.; Loike, J.; Means, T.K.; Moestrup, S.K.; et al. Standardizing scavenger receptor nomenclature. J. Immunol. 2014, 192, 1997–2006. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Lee, J.O. Structural biology of the Toll-like receptor family. Annu Rev. Biochem. 2011, 80, 917–941. [Google Scholar] [CrossRef] [PubMed]
- Gowen, B.B.; Borg, T.K.; Ghaffar, A.; Mayer, E.P. The collagenous domain of class A scavenger receptors is involved in macrophage adhesion to collagens. J. Leukoc. Biol. 2001, 69, 575–582. [Google Scholar] [PubMed]
- Mietus-Snyder, M.; Friera, A.; Glass, C.K.; Pitas, R.E. Regulation of scavenger receptor expression in smooth muscle cells by protein kinase C: a role for oxidative stress. Arterioscler Thromb. Vasc. Biol. 1997, 17, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Pitas, R.E. Expression of the acetyl low density lipoprotein receptor by rabbit fibroblasts and smooth muscle cells. Up-regulation by phorbol esters. J. Biol. Chem. 1990, 265, 12722–12727. [Google Scholar] [PubMed]
- Han, H.J.; Tokino, T.; Nakamura, Y. CSR, a scavenger receptor-like protein with a protective role against cellular damage causedby UV irradiation and oxidative stress. Hum. Mol. Genet. 1998, 7, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Paragas, N.; Ned, R.M.; Qiu, A.; Viltard, M.; Leete, T.; Drexler, I.R.; Chen, X.; Sanna-Cherchi, S.; Mohammed, F.; et al. Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev. Cell. 2009, 16, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Kangas, M.; Brannstrom, A.; Elomaa, O.; Matsuda, Y.; Eddy, R.; Shows, T.B.; Tryggvason, K. Structure and chromosomal localization of the human and murine genes for the macrophage MARCO receptor. Genomics 1999, 58, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Ojala, J.R.; Pikkarainen, T.; Tuuttila, A.; Sandalova, T.; Tryggvason, K. Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J. Biol. Chem. 2007, 282, 16654–16666. [Google Scholar] [CrossRef] [PubMed]
- Kraal, G.; van der Laan, L.J.; Elomaa, O.; Tryggvason, K. The macrophage receptor MARCO. Microbes Infect. 2000, 2, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Thelen, T.; Hao, Y.; Medeiros, A.I.; Curtis, J.L.; Serezani, C.H.; Kobzik, L.; Harris, L.H.; Aronoff, D.M. The class A scavenger receptor, macrophage receptor with collagenous structure, is the major phagocytic receptor for Clostridium sordellii expressed by human decidual macrophages. J. Immunol. 2010, 185, 4328–4335. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X.; Ben, J.; Yue, S.; Bai, H.; Guan, X.; Bai, X.; Jiang, L.; Ji, Y.; Fan, L.; et al. The di-leucine motif contributes to class a scavenger receptor-mediated internalization of acetylated lipoproteins. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.D.; Zhuang, Y.; Ben, J.J.; Qian, L.L.; Huang, H.P.; Bai, H.; Sha, J.H.; He, Z.G.; Chen, Q. Caveolae-dependent endocytosis is required for class A macrophage scavenger receptor-mediated apoptosis in macrophages. J. Biol. Chem. 2011, 286, 8231–8239. [Google Scholar] [CrossRef] [PubMed]
- Amiel, E.; Nicholson-Dykstra, S.; Walters, J.J.; Higgs, H.; Berwin, B. Scavenger receptor-A functions in phagocytosis of E. coli by bone marrow dendritic cells. Exp. Cell. Res. 2007, 313, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Kurihara, Y.; Takeya, M.; Kamada, N.; Kataoka, M.; Jishage, K.; Ueda, O.; Sakaguchi, H.; Higashi, T.; Suzuki, T.; et al. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature 1997, 386, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Kodama, T.; Suzuki, H. Macrophage scavenger receptor (SR-A I/II) deficiency reduced diet-induced atherosclerosis in C57BL/6J mice. J. Atheroscler. Thromb. 2001, 8. [Google Scholar] [CrossRef]
- Manning-Tobin, J.J.; Moore, K.J.; Seimon, T.A.; Bell, S.A.; Sharuk, M.; Alvarez-Leite, J.I.; de Winther, M.P.; Tabas, I.; Freeman, M.W. Loss of SR-A and CD36 activity reduces atherosclerotic lesion complexity without abrogating foam cell formation in hyperlipidemic mice. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Makinen, P.I.; Lappalainen, J.P.; Heinonen, S.E.; Leppanen, P.; Lahteenvuo, M.T.; Aarnio, J.V.; Heikkila, J.; Turunen, M.P.; Yla-Herttuala, S. Silencing of either SR-A or CD36 reduces atherosclerosis in hyperlipidaemic mice and reveals reciprocal upregulation of these receptors. Cardiovasc. Res. 2010, 88, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Jalkanen, J.; Leppanen, P.; Narvanen, O.; Greaves, D.R.; Yla-Herttuala, S. Adenovirus-mediated gene transfer of a secreted decoy human macrophage scavenger receptor (SR-AI) in LDL receptor knock-out mice. Atherosclerosis 2003, 169, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Ricci, R.; Sumara, G.; Sumara, I.; Rozenberg, I.; Kurrer, M.; Akhmedov, A.; Hersberger, M.; Eriksson, U.; Eberli, F.R.; Becher, B.; et al. Requirement of JNK2 for scavenger receptor A-mediated foam cell formation in atherogenesis. Science 2004, 306, 1558–1561. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, K.; Komohara, Y.; Fujiwara, Y.; Takemura, K.; Lei, X.; Nakagawa, T.; Sakashita, N.; Takeya, M. Suppression of TLR4-mediated inflammatory response by macrophage class A scavenger receptor (CD204). Biochem. Biophys. Res. Commun. 2011, 411, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.; Bauer, A.K.; Arredouani, M.; Soininen, R.; Tryggvason, K.; Kleeberger, S.R.; Kobzik, L. Protection against inhaled oxidants through scavenging of oxidized lipids by macrophage receptors MARCO and SR-AI/II. J. Clin. Invest. 2007, 117, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Varin, A.; Chen, Y.; Liu, B.; Tryggvason, K.; Gordon, S. SR-A/MARCO-mediated ligand delivery enhances intracellular TLR and NLR function, but ligand scavenging from cell surface limits TLR4 response to pathogens. Blood 2011, 117, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Jozefowski, S.; Arredouani, M.; Sulahian, T.; Kobzik, L. Disparate regulation and function of the class A scavenger receptors SR-AI/II and MARCO. J. Immunol. 2005, 175, 8032–8041. [Google Scholar] [CrossRef] [PubMed]
- Pikkarainen, T.; Brannstrom, A.; Tryggvason, K. Expression of macrophage MARCO receptor induces formation of dendritic plasma membrane processes. J. Biol. Chem. 1999, 274, 10975–10982. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pikkarainen, T.; Elomaa, O.; Soininen, R.; Kodama, T.; Kraal, G.; Tryggvason, K. Defective microarchitecture of the spleen marginal zone and impaired response to a thymus-independent type 2 antigen in mice lacking scavenger receptors MARCO and SR-A. J. Immunol. 2005, 175, 8173–8180. [Google Scholar] [CrossRef] [PubMed]
- Berwin, B.; Hart, J.P.; Rice, S.; Gass, C.; Pizzo, S.V.; Post, S.R.; Nicchitta, C.V. Scavenger receptor-A mediates gp96/GRP94 and calreticulin internalization by antigen-presenting cells. EMBO J. 2003, 22, 6127–6136. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.; Haug, M.; Kwok, W.W.; Kalbacher, H.; Wernet, D.; Dannecker, G.E.; Holzer, U. Involvement of CD91 and scavenger receptors in Hsp70-facilitated activation of human antigen-specific CD4+ memory T cells. Eur J. Immunol. 2010, 40, 986–997. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.A.; Antonopoulos, A.; Hitchen, P.G.; Haslam, S.M.; Dell, A.; Drickamer, K.; Taylor, M.E. Identification of neutrophil granule glycoproteins as Lewis(x)-containing ligands cleared by the scavenger receptor C-type lectin. J. Biol. Chem. 2011, 286, 24336–24349. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Tsuruta, Y.; Iwasaki, M.; Yamane, S.; Ochi, T.; Suzuki, R. SRCL/CL-P1 recognizes GalNAc and a carcinoma-associated antigen, Tn antigen. J. Biochem. 2003, 133, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Ohtani, K.; Fukuoh, A.; Yoshizaki, T.; Fukuda, M.; Motomura, W.; Mori, K.; Fukuzawa, J.; Kitamoto, N.; Yoshida, I.; et al. Scavenger receptor collectin placenta 1 (CL-P1) predominantly mediates zymosan phagocytosis by human vascular endothelial cells. J. Biol. Chem. 2009, 284, 3956–3965. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, K.; Suzuki, Y.; Eda, S.; Kawai, T.; Kase, T.; Keshi, H.; Sakai, Y.; Fukuoh, A.; Sakamoto, T.; Itabe, H.; et al. The membrane-type collectin CL-P1 is a scavenger receptor on vascular endothelial cells. J. Biol. Chem. 2001, 276, 44222–44228. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zheng, D.L.; Qin, F.S.; Cheng, N.; Chen, H.; Wan, B.B.; Wang, Y.P.; Xiao, H.S.; Han, Z.G. Genetic and epigenetic silencing of SCARA5 may contribute to human hepatocellular carcinoma by activating FAK signaling. J. Clin. Invest. 2010, 120, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Zhang, S.; Yang, Y.; Cheng, L.; Li, C.; Dai, L.; Zhang, X.; Fan, P.; Tian, H.; Wang, R.; et al. Therapeutic upregulation of SCARA5 inhibits tumor growth and metastasis. Cancer Sci. 2012, 103, 1631–1639. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Oliver, P.; Davies, K.E.; Platt, N. Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. J. Biol. Chem. 2006, 281, 11834–11845. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Chen, M.; Song, Z.; Daugherty, A.; Li, X.A. C323 of SR-BI is required for SR-BI-mediated HDL binding and cholesteryl ester uptake. J. Lipid Res. 2011, 52, 2272–2278. [Google Scholar] [CrossRef] [PubMed]
- Scarselli, E.; Ansuini, H.; Cerino, R.; Roccasecca, R.M.; Acali, S.; Filocamo, G.; Traboni, C.; Nicosia, A.; Cortese, R.; Vitelli, A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002, 21, 5017–5025. [Google Scholar] [CrossRef] [PubMed]
- Yates, M.; Kolmakova, A.; Zhao, Y.; Rodriguez, A. Clinical impact of scavenger receptor class B type I gene polymorphisms on human female fertility. Hum. Reprod 2011, 26, 1910–1916. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, Y.; Kimura-Matsumoto, M.; Murakami, M.; Yamamoto, K.; Akasaka, Y.; Uzuki, M.; Yuri, Y.; Inomata, N.; Yokoo, T.; Ishii, T. Distribution of smooth muscle cells and macrophages expressing scavenger receptor BI/II in atherosclerosis. J. Atheroscler. Thromb. 2009, 16, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, E.R.; Cai, L.; Sun, B.; Webb, N.R.; van der Westhuyzen, D.R. High density lipoprotein uptake by scavenger receptor SR-BII. J. Biol. Chem. 2004, 279, 14372–14381. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, R.L.; Li, W.; Park, Y.M.; Rahaman, S.O. Mechanisms of cell signaling by the scavenger receptor CD36: Implications in atherosclerosis and thrombosis. Trans. Am. Clin. Climatol. Assoc. 2010, 121, 206–220. [Google Scholar] [PubMed]
- Liani, R.; Halvorsen, B.; Sestili, S.; Handberg, A.; Santilli, F.; Vazzana, N.; Formoso, G.; Aukrust, P.; Davi, G. Plasma levels of soluble CD36, platelet activation, inflammation, and oxidative stress are increased in type 2 diabetic patients. Free Radic. Biol. Med. 2012, 52, 1318–1324. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, S.; Sugimoto, J.; Okada, M.; Sakairi, T.; Takagi, S. Gene expression in livers of BALB/C and C57BL/6J mice fed a high-fat diet. Toxicol. Pathol. 2012, 40, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, R.L.; Febbraio, M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009, 2, re3. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M.; Drazba, J.A.; Vasanji, A.; Egelhoff, T.; Febbraio, M.; Silverstein, R.L. Oxidized LDL/CD36 interaction induces loss of cell polarity and inhibits macrophage locomotion. Mol. Biol. Cell. 2012, 23, 3057–3068. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.R.; Stuart, L.M.; Wilkinson, K.; van Gils, J.M.; Deng, J.; Halle, A.; Rayner, K.J.; Boyer, L.; Zhong, R.; Frazier, W.A.; et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 2010, 11, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Gowda, N.M.; Wu, X.; Gowda, R.N.; Gowda, D.C. CD36 modulates proinflammatory cytokine responses to Plasmodium falciparum glycosylphosphatidylinositols and merozoites by dendritic cells. Parasite Immunol. 2012, 34, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Acton, S.L.; Scherer, P.E.; Lodish, H.F.; Krieger, M. Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J. Biol. Chem. 1994, 269, 21003–21009. [Google Scholar] [PubMed]
- Nieland, T.J.; Ehrlich, M.; Krieger, M.; Kirchhausen, T. Endocytosis is not required for the selective lipid uptake mediated by murine SR-BI. Biochim Biophys Acta 2005, 1734, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Boyanovsky, B.B.; Connelly, M.A.; Shridas, P.; van der Westhuyzen, D.R.; Webb, N.R. Distinct mechanisms for OxLDL uptake and cellular trafficking by class B scavenger receptors CD36 and SR-BI. J. Lipid Res. 2007, 48, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Tancevski, I.; Frank, S.; Massoner, P.; Stanzl, U.; Schgoer, W.; Wehinger, A.; Fievet, C.; Eller, P.; Patsch, J.R.; Ritsch, A. Increased plasma levels of LDL cholesterol in rabbits after adenoviral overexpression of human scavenger receptor class B type I. J. Mol. Med. (Berl) 2005, 83, 927–932. [Google Scholar] [CrossRef]
- Peng, Y.; Akmentin, W.; Connelly, M.A.; Lund-Katz, S.; Phillips, M.C.; Williams, D.L. Scavenger receptor BI (SR-BI) clustered on microvillar extensions suggests that this plasma membrane domain is a way station for cholesterol trafficking between cells and high-density lipoprotein. Mol. Biol. Cell. 2004, 15, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Yamayoshi, S.; Fujii, K.; Koike, S. Scavenger receptor b2 as a receptor for hand, foot, and mouth disease and severe neurological diseases. Front. Microbiol. 2012, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- Yamayoshi, S.; Iizuka, S.; Yamashita, T.; Minagawa, H.; Mizuta, K.; Okamoto, M.; Nishimura, H.; Sanjoh, K.; Katsushima, N.; Itagaki, T.; et al. Human SCARB2-dependent infection by coxsackievirus A7, A14, and A16 and enterovirus 71. J. Virol. 2012, 86, 5686–5696. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, E.R.; Cai, L.; Shetty, S.; Zhao, Z.; Szanto, A.; Webb, N.R.; Van der Westhuyzen, D.R. High density lipoprotein endocytosis by scavenger receptor SR-BII is clathrin-dependent and requires a carboxyl-terminal dileucine motif. J. Biol. Chem. 2006, 281, 4348–4353. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Marin, E.; Fernandez-Prieto, L.; Rodriguez-Del Rio, E.; Madrazo-Toca, F.; Reinheckel, T.; Saftig, P.; Alvarez-Dominguez, C. LIMP-2 links late phagosomal trafficking with the onset of the innate immune response to Listeria monocytogenes: A role in macrophage activation. J. Biol. Chem. 2011, 286, 3332–3341. [Google Scholar] [CrossRef] [PubMed]
- Innate Immunity; Ezekowitz, R.A.B.; Hoffman, J.A. (Eds.) Humana press: Totowa, NJ, USA, 2003.
- Ramet, M.; Pearson, A.; Manfruelli, P.; Li, X.; Koziel, H.; Gobel, V.; Chung, E.; Krieger, M.; Ezekowitz, R.A. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity 2001, 15, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Li, A.C.; Guidez, F.R.; Collier, J.G.; Glass, C.K. The macrosialin promoter directs high levels of transcriptional activity in macrophages dependent on combinatorial interactions between PU.1 and c-Jun. J. Biol. Chem. 1998, 273, 5389–5399. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Quehenberger, O.; Kondratenko, N.; Green, S.; Steinberg, D. Minimally oxidized low-density lipoprotein increases expression of scavenger receptor A, CD36, and macrosialin in resident mouse peritoneal macrophages. Arterioscler. Thromb. Vasc. Biol. 1998, 18, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Holness, C.L.; Simmons, D.L. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood 1993, 81, 1607–1613. [Google Scholar] [PubMed]
- Ramprasad, M.P.; Terpstra, V.; Kondratenko, N.; Quehenberger, O.; Steinberg, D. Cell surface expression of mouse macrosialin and human CD68 and their role as macrophage receptors for oxidized low density lipoprotein. Proc. Natl. Acad. Sci. USA 1996, 93, 14833–14838. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.P.; Gordon, S. Phagocytosis stimulates alternative glycosylation of macrosialin (mouse CD68), a macrophage-specific endosomal protein. Biochem. J. 1999, 338, 687–694. [Google Scholar] [PubMed]
- Zeibig, S.; Li, Z.; Wagner, S.; Holthoff, H.P.; Ungerer, M.; Bultmann, A.; Uhland, K.; Vogelmann, J.; Simmet, T.; Gawaz, M.; et al. Effect of the oxLDL binding protein Fc-CD68 on plaque extension and vulnerability in atherosclerosis. Circ. Res. 2011, 108, 695–703. [Google Scholar] [CrossRef] [PubMed]
- De Beer, M.C.; Zhao, Z.; Webb, N.R.; van der Westhuyzen, D.R.; de Villiers, W.J. Lack of a direct role for macrosialin in oxidized LDL metabolism. J. Lipid Res. 2003, 44, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Lee, C.; Schindler, C. Deletion of the murine scavenger receptor CD68. J. Lipid Res. 2011, 52, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Goyal, T.; Mehta, J.L. Oxidized LDL, LOX-1 and atherosclerosis. Cardiovasc. Drugs Ther. 2011, 25, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sawamura, T.; Kurdowska, A.K.; Ji, H.L.; Idell, S.; Fu, J. LOX-1 deletion improves neutrophil responses, enhances bacterial clearance, and reduces lung injury in a murine polymicrobial sepsis model. Infect. Immun. 2011, 79, 2865–2870. [Google Scholar] [CrossRef] [PubMed]
- Mehta, J.L.; Sanada, N.; Hu, C.P.; Chen, J.; Dandapat, A.; Sugawara, F.; Satoh, H.; Inoue, K.; Kawase, Y.; Jishage, K.; et al. Deletion of LOX-1 reduces atherogenesis in LDLR knockout mice fed high cholesterol diet. Circ. Res. 2007, 100, 1634–1642. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, L.; Chen, H.; Sawamura, T.; Ranganathan, S.; Mehta, J.L. LOX-1 mediates oxidized low-density lipoprotein-induced expression of matrix metalloproteinases in human coronary artery endothelial cells. Circulation 2003, 107, 612–617. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Arai, Y.; Kurihara, H.; Kita, T.; Sawamura, T. Overexpression of lectin-like oxidized low-density lipoprotein receptor-1 induces intramyocardial vasculopathy in apolipoprotein E-null mice. Circ. Res. 2005, 97, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Ishigaki, Y.; Katagiri, H.; Gao, J.; Yamada, T.; Imai, J.; Uno, K.; Hasegawa, Y.; Kaneko, K.; Ogihara, T.; Ishihara, H.; et al. Impact of plasma oxidized low-density lipoprotein removal on atherosclerosis. Circulation 2008, 118, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Kume, N.; Kita, T. Lectin-like oxidized low-density lipoprotein receptor-1 (LOX-1) in atherogenesis. Trends Cardiovasc Med. 2001, 11, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, H.; Kume, N.; Miyamoto, S.; Minami, M.; Morimoto, M.; Hayashida, K.; Hashimoto, N.; Kita, T. Oxidized LDL modulates Bax/Bcl-2 through the lectinlike Ox-LDL receptor-1 in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Biocca, S.; Falconi, M.; Filesi, I.; Baldini, F.; Vecchione, L.; Mango, R.; Romeo, F.; Federici, G.; Desideri, A.; Novelli, G. Functional analysis and molecular dynamics simulation of LOX-1 K167N polymorphism reveal alteration of receptor activity. PLoS ONE 2009, 4, e4648. [Google Scholar] [CrossRef] [PubMed]
- Khaidakov, M.; Wang, X.; Mehta, J.L. Potential involvement of LOX-1 in functional consequences of endothelial senescence. PLoS ONE 2011, 6, e20964. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, T.; Kume, N.; Minami, M.; Hayashida, K.; Sawamura, T.; Kita, T.; Yonehara, S. LOX-1 supports adhesion of Gram-positive and Gram-negative bacteria. J. Immunol. 2001, 166, 5108–5114. [Google Scholar] [CrossRef] [PubMed]
- Kielian, T.; Haney, A.; Mayes, P.M.; Garg, S.; Esen, N. Toll-like receptor 2 modulates the proinflammatory milieu in Staphylococcus aureus-induced brain abscess. Infect. Immun. 2005, 73, 7428–7435. [Google Scholar] [CrossRef] [PubMed]
- Jeannin, P.; Bottazzi, B.; Sironi, M.; Doni, A.; Rusnati, M.; Presta, M.; Maina, V.; Magistrelli, G.; Haeuw, J.F.; Hoeffel, G.; et al. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity 2005, 22, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Tsujimoto, M. Characterization of the human gene encoding the scavenger receptor expressed by endothelial cell and its regulation by a novel transcription factor, endothelial zinc finger protein-2. J. Biol. Chem. 2002, 277, 24014–24021. [Google Scholar] [CrossRef] [PubMed]
- Ishii, J.; Adachi, H.; Aoki, J.; Koizumi, H.; Tomita, S.; Suzuki, T.; Tsujimoto, M.; Inoue, K.; Arai, H. SREC-II, a new member of the scavenger receptor type F family, trans-interacts with SREC-I through its extracellular domain. J. Biol. Chem. 2002, 277, 39696–39702. [Google Scholar] [CrossRef] [PubMed]
- Murshid, A.; Gong, J.; Calderwood, S.K. Heat shock protein 90 mediates efficient antigen cross presentation through the scavenger receptor expressed by endothelial cells-I. J. Immunol. 2010, 185, 2903–2917. [Google Scholar] [CrossRef] [PubMed]
- Sano, M.; Korekane, H.; Ohtsubo, K.; Yamaguchi, Y.; Kato, M.; Shibukawa, Y.; Tajiri, M.; Adachi, H.; Wada, Y.; Asahi, M.; et al. N-glycans of SREC-I (scavenger receptor expressed by endothelial cells): essential role for ligand binding, trafficking and stability. Glycobiology 2012, 22, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Gutwein, P.; Abdel-Bakky, M.S.; Schramme, A.; Doberstein, K.; Kampfer-Kolb, N.; Amann, K.; Hauser, I.A.; Obermuller, N.; Bartel, C.; Abdel-Aziz, A.A.; et al. CXCL16 is expressed in podocytes and acts as a scavenger receptor for oxidized low-density lipoprotein. Am. J. Pathol 2009, 174, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Gursel, M.; Gursel, I.; Mostowski, H.S.; Klinman, D.M. CXCL16 influences the nature and specificity of CpG-induced immune activation. J. Immunol. 2006, 177, 1575–1580. [Google Scholar] [CrossRef] [PubMed]
- Sheikine, Y.; Sirsjo, A. CXCL16/SR-PSOX—A friend or a foe in atherosclerosis? Atherosclerosis 2008, 197, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, R.; Tanaka, M.; Kume, N.; Minami, M.; Kawamoto, T.; Togi, K.; Shimaoka, T.; Takahashi, S.; Yamaguchi, J.; Nishina, T.; et al. Upregulation of SR-PSOX/CXCL16 and recruitment of CD8+ T cells in cardiac valves during inflammatory valvular heart disease. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Wagsater, D.; Olofsson, P.S.; Norgren, L.; Stenberg, B.; Sirsjo, A. The chemokine and scavenger receptor CXCL16/SR-PSOX is expressed in human vascular smooth muscle cells and is induced by interferon gamma. Biochem. Biophys. Res. Commun. 2004, 325, 1187–1193. [Google Scholar] [CrossRef] [PubMed]
- Wuttge, D.M.; Zhou, X.; Sheikine, Y.; Wagsater, D.; Stemme, V.; Hedin, U.; Stemme, S.; Hansson, G.K.; Sirsjo, A. CXCL16/SR-PSOX is an interferon-gamma-regulated chemokine and scavenger receptor expressed in atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, H.J.; Li, T.J.; Yang, Y.; Guo, X.L.; Wu, M.C.; Rui, Y.C.; Wei, L.X. Lentiviral vector-mediated siRNA knockdown of SR-PSOX inhibits foam cell formation in vitro. Acta Pharmacol. Sin. 2008, 29, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Quan, Z.; Yang, H.; Yang, Y.; Yan, B.; Cao, R.; Wen, G.; Liu, C.; Xu, Y. Construction and functional analysis of a lentiviral expression vector containing a scavenger receptor (SR-PSOX) that binds uniquely phosphatidylserine and oxidized lipoprotein. Acta Biochim. Biophys. Sin. 2007, 39, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, G.A.; Kellin, A.; Samnegard, A.; Lundman, P.; Tornvall, P.; Dimmeler, S.; Zeiher, A.M.; Hamsten, A.; Hansson, G.K.; Eriksson, P. Severity of coronary artery stenosis is associated with a polymorphism in the CXCL16/SR-PSOX gene. J. Intern. Med. 2005, 257, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Xie, C.; Wang, H.W.; Zhou, X.J.; Schwartz, N.; Calixto, S.; Mackay, M.; Aranow, C.; Putterman, C.; Mohan, C. Elevated urinary VCAM-1, P-selectin, soluble TNF receptor-1, and CXC chemokine ligand 16 in multiple murine lupus strains and human lupus nephritis. J. Immunol. 2007, 179, 7166–7175. [Google Scholar] [CrossRef] [PubMed]
- Uza, N.; Nakase, H.; Yamamoto, S.; Yoshino, T.; Takeda, Y.; Ueno, S.; Inoue, S.; Mikami, S.; Matsuura, M.; Shimaoka, T.; et al. SR-PSOX/CXCL16 plays a critical role in the progression of colonic inflammation. Gut 2011, 60, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, T.; Seino, K.; Kume, N.; Minami, M.; Nishime, C.; Suematsu, M.; Kita, T.; Taniguchi, M.; Matsushima, K.; Yonehara, S. Critical role for CXC chemokine ligand 16 (SR-PSOX) in Th1 response mediated by NKT cells. J. Immunol. 2007, 179, 8172–8179. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, T.; Nakayama, T.; Fukumoto, N.; Kume, N.; Takahashi, S.; Yamaguchi, J.; Minami, M.; Hayashida, K.; Kita, T.; Ohsumi, J.; et al. Cell surface-anchored SR-PSOX/CXC chemokine ligand 16 mediates firm adhesion of CXC chemokine receptor 6-expressing cells. J. Leukoc. Biol. 2004, 75, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Gratchev, A.; Sobenin, I.; Orekhov, A.; Kzhyshkowska, J. Monocytes as a diagnostic marker of cardiovascular diseases. Immunobiology 2012, 217, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Tsujimoto, M. FEEL-1, a novel scavenger receptor with in vitro bacteria-binding and angiogenesis-modulating activities. J. Biol. Chem. 2002, 277, 34264–34270. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.; Weston, C.J.; Oo, Y.H.; Westerlund, N.; Stamataki, Z.; Youster, J.; Hubscher, S.G.; Salmi, M.; Jalkanen, S.; Lalor, P.F.; et al. Common lymphatic endothelial and vascular endothelial receptor-1 mediates the transmigration of regulatory T cells across human hepatic sinusoidal endothelium. J. Immunol. 2011, 186, 4147–4155. [Google Scholar] [CrossRef] [PubMed]
- Palani, S.; Maksimow, M.; Miiluniemi, M.; Auvinen, K.; Jalkanen, S.; Salmi, M. Stabilin-1/CLEVER-1, a type 2 macrophage marker, is an adhesion and scavenging molecule on human placental macrophages. Eur. J. Immunol. 2011, 41, 2052–2063. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Tsujimoto, M. Adaptor protein sorting nexin 17 interacts with the scavenger receptor FEEL-1/stabilin-1 and modulates its expression on the cell surface. Biochim. Biophys. Acta 2010, 1803, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Kzhyshkowska, J.; Workman, G.; Cardo-Vila, M.; Arap, W.; Pasqualini, R.; Gratchev, A.; Krusell, L.; Goerdt, S.; Sage, E.H. Novel function of alternatively activated macrophages: stabilin-1-mediated clearance of SPARC. J. Immunol. 2006, 176, 5825–5832. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kim, S.Y.; Kang, K.B.; Kim, I.S. Adaptor protein GULP is involved in stabilin-1-mediated phagocytosis. Biochem. Biophys. Res. Commun. 2010, 398, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, S.Y.; Kim, S.Y.; Bae, D.J.; Pyo, J.H.; Hong, M.; Kim, I.S. Cross Talk between Engulfment Receptors Stabilin-2 and Integrin alphavbeta5 Orchestrates Engulfment of Phosphatidylserine-Exposed Erythrocytes. Mol. Cell. Biol. 2012, 32, 2698–2708. [Google Scholar] [CrossRef] [PubMed]
- Moeller, J.B.; Nielsen, M.J.; Reichhardt, M.P.; Schlosser, A.; Sorensen, G.L.; Nielsen, O.; Tornoe, I.; Gronlund, J.; Nielsen, M.E.; Jorgensen, J.S.; et al. CD163-L1 is an endocytic macrophage protein strongly regulated by mediators in the inflammatory response. J. Immunol. 2012, 188, 2399–2409. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.J.; Madsen, M.; Moller, H.J.; Moestrup, S.K. The macrophage scavenger receptor CD163: Endocytic properties of cytoplasmic tail variants. J. Leukoc. Biol. 2006, 79, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, M.; Graversen, J.H.; Jacobsen, C.; Sonne, O.; Hoffman, H.J.; Law, S.K.; Moestrup, S.K. Identification of the haemoglobin scavenger receptor. Nature 2001, 409, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, J.H.; Etzerodt, A.; Svendsen, P.; Moestrup, S.K. The haptoglobin-CD163-heme oxygenase-1 pathway for hemoglobin scavenging. Oxid Med. Cell. Longev. 2013, 2013. [Google Scholar] [CrossRef]
- Canton, J.; Neculai, D.; Grinstein, S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013, 13, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Alonso, R.; Buors, C.; Le Dantec, C.; Hillion, S.; Pers, J.O.; Saraux, A.; Montero, E.; Marianowski, R.; Loisel, S.; Devauchelle, V.; et al. Aberrant expression of CD6 on B-cell subsets from patients with Sjogren's syndrome. J. Autoimmun. 2010, 35, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Burdo, T.H.; Lentz, M.R.; Autissier, P.; Krishnan, A.; Halpern, E.; Letendre, S.; Rosenberg, E.S.; Ellis, R.J.; Williams, K.C. Soluble CD163 made by monocyte/macrophages is a novel marker of HIV activity in early and chronic infection prior to and after anti-retroviral therapy. J. Infect. Dis. 2011, 204, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Etzerodt, A.; Moestrup, S.K. CD163 and inflammation: biological, diagnostic, and therapeutic aspects. Antioxid. Redox Signal. 2013, 18, 2352–2363. [Google Scholar] [CrossRef] [PubMed]
- Fabriek, B.O.; Polfliet, M.M.; Vloet, R.P.; van der Schors, R.C.; Ligtenberg, A.J.; Weaver, L.K.; Geest, C.; Matsuno, K.; Moestrup, S.K.; Dijkstra, C.D.; et al. The macrophage CD163 surface glycoprotein is an erythroblast adhesion receptor. Blood 2007, 109, 5223–5229. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, R.; Yan, S.F.; Schmidt, A.M. RAGE: therapeutic target and biomarker of the inflammatory response--the evidence mounts. J. Leukoc. Biol. 2009, 86, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.D.; Chen, X.; Fu, J.; Chen, M.; Zhu, H.; Roher, A.; Slattery, T.; Zhao, L.; Nagashima, M.; Morser, J.; et al. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer’s disease. Nature 1996, 382, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.A.; Drury, S.; Fu, C.; Qu, W.; Taguchi, A.; Lu, Y.; Avila, C.; Kambham, N.; Bierhaus, A.; Nawroth, P.; et al. RAGE mediates a novel proinflammatory axis: A central cell surface receptor for S100/calgranulin polypeptides. Cell 1999, 97, 889–901. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Kubo, H.; Morimoto, K.; Fujino, N.; Suzuki, T.; Takahasi, T.; Yamada, M.; Yamaya, M.; Maekawa, T.; Yamamoto, Y.; et al. Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep. 2011, 12, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Hori, O.; Brett, J.; Slattery, T.; Cao, R.; Zhang, J.; Chen, J.X.; Nagashima, M.; Lundh, E.R.; Vijay, S.; Nitecki, D.; et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J. Biol. Chem. 1995, 270, 25752–25761. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Schmidt, A.M. Characterization and functional analysis of the promoter of RAGE, the receptor for advanced glycation end products. J. Biol. Chem. 1997, 272, 16498–16506. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Brownlee, M. Hyperglycemia-induced reactive oxygen species increase expression of the receptor for advanced glycation end products (RAGE) and RAGE ligands. Diabetes 2010, 59, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Mendez, J.D.; Mendez-Valenzuela, V.; Aguilar-Hernandez, M.M. Cellular signalling of the receptor for advanced glycation end products (RAGE). Cell. Signal. 2013, 25, 2185–2197. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Sturgis, L.; Haidacher, J.; Zhang, X.N.; Sherwood, S.J.; Bjercke, R.J.; Juhasz, O.; Crow, M.T.; Tilton, R.G.; Denner, L. Requirement for p38 and p44/p42 mitogen-activated protein kinases in RAGE-mediated nuclear factor-kappaB transcriptional activation and cytokine secretion. Diabetes 2001, 50, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Harja, E.; Bu, D.X.; Hudson, B.I.; Chang, J.S.; Shen, X.; Hallam, K.; Kalea, A.Z.; Lu, Y.; Rosario, R.H.; Oruganti, S.; et al. Vascular and inflammatory stresses mediate atherosclerosis via RAGE and its ligands in apoE-/- mice. J. Clin. Invest. 2008, 118, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chen, X. Ox-LDL-induced LOX-1 expression in vascular smooth muscle cells: role of reactive oxygen species. Fundam. Clin. Pharmacol. 2011, 25, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Cominacini, L.; Fratta Pasini, A.; Garbin, U.; Pastorino, A.; Rigoni, A.; Nava, C.; Davoli, A.; Lo Cascio, V.; Sawamura, T. The platelet-endothelium interaction mediated by lectin-like oxidized low-density lipoprotein receptor-1 reduces the intracellular concentration of nitric oxide in endothelial cells. J. Am. Coll. Cardiol. 2003, 41, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Dandapat, A.; Chen, J.; Fujita, Y.; Inoue, N.; Kawase, Y.; Jishage, K.; Suzuki, H.; Sawamura, T.; Mehta, J.L. LOX-1 deletion alters signals of myocardial remodeling immediately after ischemia-reperfusion. Cardiovasc. Res. 2007, 76, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Hokari, S.; Koyama, I.; Harada, T.; Komoda, T. NF-kappa B activation in endothelial cells treated with oxidized high-density lipoprotein. Biochem. Biophys. Res. Commun. 2003, 303, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Gaytan, R.; Mas-Oliva, J. Oxidative stress impairs endocytosis of the scavenger receptor class A. Biochem. Biophys. Res. Commun. 2003, 305, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Febbraio, M.; Reddy, S.P.; Yu, D.Y.; Yamamoto, M.; Silverstein, R.L. CD36 participates in a signaling pathway that regulates ROS formation in murine VSMCs. J. Clin. Invest. 2010, 120, 3996–4006. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Park, E.M.; Febbraio, M.; Anrather, J.; Park, L.; Racchumi, G.; Silverstein, R.L.; Iadecola, C. The class B scavenger receptor CD36 mediates free radical production and tissue injury in cerebral ischemia. J. Neurosci. 2005, 25, 2504–2512. [Google Scholar] [CrossRef] [PubMed]
- Boullier, A.; Bird, D.A.; Chang, M.K.; Dennis, E.A.; Friedman, P.; Gillotre-Taylor, K.; Horkko, S.; Palinski, W.; Quehenberger, O.; Shaw, P.; et al. Scavenger receptors, oxidized LDL, and atherosclerosis. Ann. N. Y. Acad. Sci. 2001, 947, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Lehtolainen, P.; Takeya, M.; Yla-Herttuala, S. Retrovirus-mediated, stable scavenger-receptor gene transfer leads to functional endocytotic receptor expression, foam cell formation, and increased susceptibility to apoptosis in rabbit aortic smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, R.F., Jr.; Thakur, S.A.; Mayfair, J.K.; Holian, A. MARCO mediates silica uptake and toxicity in alveolar macrophages from C57BL/6 mice. J. Biol. Chem. 2006, 281, 34218–34226. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.H.; Yu, Y.P.; Shi, Y.K.; Nelson, J.B.; Luo, J.H. CSR1 induces cell death through inactivation of CPSF3. Oncogene 2009, 28, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Nishiuchi, T.; Murao, K.; Imachi, H.; Yu, X.; Dobashi, H.; Haba, R.; Ishida, T. Scavenger receptor class BI mediates the anti-apoptotic effect of erythropoietin. Ann. Med. 2010, 42, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Yang, M.; Yuen, P.M.; Chik, K.W.; Li, C.K.; Shing, M.M.; Lam, H.K.; Fok, T.F. Thrombospondin-1 induces apoptosis in primary leukemia and cell lines mediated by CD36 and Caspase-3. Int. J. Mol. Med. 2003, 12, 995–1001. [Google Scholar] [PubMed]
- Lu, J.; Yang, J.H.; Burns, A.R.; Chen, H.H.; Tang, D.; Walterscheid, J.P.; Suzuki, S.; Yang, C.Y.; Sawamura, T.; Chen, C.H. Mediation of electronegative low-density lipoprotein signaling by LOX-1: a possible mechanism of endothelial apoptosis. Circ. Res. 2009, 104, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Mehta, J.L.; Haider, N.; Zhang, X.; Narula, J.; Li, D. Role of caspases in Ox-LDL-induced apoptotic cascade in human coronary artery endothelial cells. Circ. Res. 2004, 94, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.E.; Tacon, D.; Tedbury, P.R.; Hadden, J.M.; Knowling, S.; Sawamura, T.; Peckham, M.; Phillips, S.E.; Walker, J.H.; Ponnambalam, S. LOX-1 scavenger receptor mediates calcium-dependent recognition of phosphatidylserine and apoptotic cells. Biochem. J. 2006, 393, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Oka, K.; Sawamura, T.; Kikuta, K.; Itokawa, S.; Kume, N.; Kita, T.; Masaki, T. Lectin-like oxidized low-density lipoprotein receptor 1 mediates phagocytosis of aged/apoptotic cells in endothelial cells. Proc. Natl. Acad. Sci. USA 1998, 95, 9535–9540. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Jung, M.Y.; Kim, H.J.; Lee, S.J.; Kim, S.Y.; Lee, B.H.; Kwon, T.H.; Park, R.W.; Kim, I.S. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell. Death Differ. 2008, 15, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Jung, M.Y.; Lee, S.J.; Kang, K.B.; Gratchev, A.; Riabov, V.; Kzhyshkowska, J.; Kim, I.S. Stabilin-1 mediates phosphatidylserine-dependent clearance of cell corpses in alternatively activated macrophages. J. Cell. Sci. 2009, 122, 3365–3373. [Google Scholar] [CrossRef] [PubMed]
- Bak, S.P.; Walters, J.J.; Takeya, M.; Conejo-Garcia, J.R.; Berwin, B.L. Scavenger receptor-A-targeted leukocyte depletion inhibits peritoneal ovarian tumor progression. Cancer Res. 2007, 67, 4783–4789. [Google Scholar] [CrossRef] [PubMed]
- Neyen, C.; Pluddemann, A.; Mukhopadhyay, S.; Maniati, E.; Bossard, M.; Gordon, S.; Hagemann, T. Macrophage scavenger receptor a promotes tumor progression in murine models of ovarian and pancreatic cancer. J. Immunol. 2013, 190, 3798–3805. [Google Scholar] [CrossRef] [PubMed]
- Bock, A.J.; Nymoen, D.A.; Brenne, K.; Kaern, J.; Davidson, B. SCARA3 mRNA is overexpressed in ovarian carcinoma compared with breast carcinoma effusions. Hum. Pathol. 2012, 43, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Tseng, G.C.; Yu, Y.P.; Gavel, T.; Nelson, J.; Wells, A.; Michalopoulos, G.; Kokkinakis, D.; Luo, J.H. CSR1 suppresses tumor growth and metastasis of prostate cancer. Am. J. Pathol 2006, 168, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Shabo, I.; Stal, O.; Olsson, H.; Dore, S.; Svanvik, J. Breast cancer expression of CD163, a macrophage scavenger receptor, is related to early distant recurrence and reduced patient survival. Int. J. Cancer 2008, 123, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Shabo, I.; Olsson, H.; Sun, X.F.; Svanvik, J. Expression of the macrophage antigen CD163 in rectal cancer cells is associated with early local recurrence and reduced survival time. Int. J. Cancer 2009, 125, 1826–1831. [Google Scholar] [CrossRef] [PubMed]
- Llaverias, G.; Danilo, C.; Wang, Y.; Witkiewicz, A.K.; Daumer, K.; Lisanti, M.P.; Frank, P.G. A Western-type diet accelerates tumor progression in an autochthonous mouse model of prostate cancer. Am. J. Pathol. 2010, 177, 3180–3191. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.S.; Li, M.; Sinyuk, M.; Jahnen-Dechent, W.; Lathia, J.D.; Silverstein, R.L. Context dependent role of the CD36--thrombospondin--histidine-rich glycoprotein axis in tumor angiogenesis and growth. PLoS ONE 2012, 7, e40033. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yan, M.; Mehta, J.L.; Hu, C. Angiogenesis is a link between atherosclerosis and tumorigenesis: role of LOX-1. Cardiovasc Drugs Ther. 2011, 25, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Khaidakov, M.; Mehta, J.L. Oxidized LDL triggers pro-oncogenic signaling in human breast mammary epithelial cells partly via stimulation of MiR-21. PLoS ONE 2012, 7, e46973. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Zhang, P.; Fu, J. Up-regulation of LOX-1 expression by TNF-alpha promotes trans-endothelial migration of MDA-MB-231 breast cancer cells. Cancer Lett. 2007, 258, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Irjala, H.; Alanen, K.; Grenman, R.; Heikkila, P.; Joensuu, H.; Jalkanen, S. Mannose receptor (MR) and common lymphatic endothelial and vascular endothelial receptor (CLEVER)-1 direct the binding of cancer cells to the lymph vessel endothelium. Cancer Res. 2003, 63, 4671–4676. [Google Scholar] [PubMed]
- Facciponte, J.G.; Wang, X.Y.; Subjeck, J.R. Hsp110 and Grp170, members of the Hsp70 superfamily, bind to scavenger receptor-A and scavenger receptor expressed by endothelial cells-I. Eur. J. Immunol. 2007, 37, 2268–2279. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zani, I.A.; Stephen, S.L.; Mughal, N.A.; Russell, D.; Homer-Vanniasinkam, S.; Wheatcroft, S.B.; Ponnambalam, S. Scavenger Receptor Structure and Function in Health and Disease. Cells 2015, 4, 178-201. https://doi.org/10.3390/cells4020178
Zani IA, Stephen SL, Mughal NA, Russell D, Homer-Vanniasinkam S, Wheatcroft SB, Ponnambalam S. Scavenger Receptor Structure and Function in Health and Disease. Cells. 2015; 4(2):178-201. https://doi.org/10.3390/cells4020178
Chicago/Turabian StyleZani, Izma Abdul, Sam L. Stephen, Nadeem A. Mughal, David Russell, Shervanthi Homer-Vanniasinkam, Stephen B. Wheatcroft, and Sreenivasan Ponnambalam. 2015. "Scavenger Receptor Structure and Function in Health and Disease" Cells 4, no. 2: 178-201. https://doi.org/10.3390/cells4020178
APA StyleZani, I. A., Stephen, S. L., Mughal, N. A., Russell, D., Homer-Vanniasinkam, S., Wheatcroft, S. B., & Ponnambalam, S. (2015). Scavenger Receptor Structure and Function in Health and Disease. Cells, 4(2), 178-201. https://doi.org/10.3390/cells4020178