1. Introduction
Wheat (
Triticum aestivum L.) is one of the most important cereals in the nation and widely consumed around the world. Increases in population, changes in diet, social and policy issues especially in developing countries, will increase the demand for staple foods such as wheat [
1]. Among the challenges that researchers are facing, the most daunting is how to increase production in a sustainable way with minimum increase in area. Ray et al. [
2] using historical data determined that wheat yield is increasing by 0.9% per year, substantially lower than the rate that we need (~2.4% per year by 2050). In addition, increased temperature and changes in rainfall patterns over the next decades will require extra effort to increase grain production [
3]. Without efficient selection of adapted plants and improvement in genetic material, a global decrease in production is estimated on the order of 6.0% in wheat, 7.4% in maize, 3.2% in rice and 3.1% in soybean for each degree-Celsius increase in temperature [
4].
IPCC [
3] projects a global increase, under all emission scenarios, in air temperature by 1 to 3.7 °C by the end of this century. Decrease in crop production due to weather events is not only a future problem, in that drought, flood and extreme temperatures are already reducing production worldwide. Lesk et al. [
5] estimated that from 1964 to 2007 around 9–10% of the reduction in cereal production worldwide was due to drought and extreme heat events. They determined that yield losses due to drought were associated with decreased harvested area and heat events, though extreme heat was the primary factor in yield decreases. In a more regional analysis of data from wheat field trials in the United States from 1985 to 2013, there was a yield reduction due to extreme heat in spring [
6]. In addition, Tack et al. [
6] observed that recently released cultivars had a lower ability to resist heat stress than old varieties. Climate change is expected to increase not only air temperature but also soil temperature. Studies with winter wheat [
7] and barley [
8] testing soil warming conditions showed limited crop development and yield production.
Climate change will also affect disease occurrence, distribution and intensity. A major disease threat to wheat and other small grains is
Fusarium head blight (FHB) which causes yield losses, decreases in grain quality and toxin production [
9]. Using a modeling approach, Backhouse et al. [
10] found a positive correlation between climate and distribution of pathogenic species of
Fusarium which includes
Fusarium graminearum. His group predicted wide distribution in countries where this disease already occurs and further, that new regions such as Mexico, North Africa, Ethiopia and western Siberia would be vulnerable to FHB epidemics [
10].
Despite the ability of this pathogen to reduce yield, the production of mycotoxins such as deoxynivalenol (DON) which are harmful to plants, animal and humans is the driving force behind selection for resistance to FHB [
9]. A study predicting wheat phenology and DON in north-western Europe pointed out that due to climate change, flowering and maturity will be 1 to 2 weeks earlier in the season and DON levels will increase up to 3 times in most of the regions where the study was carried out [
11]. Fusarium damaged kernels (FDK) is also an important trait since kernel damage is associated with reduced test weight, which directly affects farmers who receive lower prices for their grain.
Success during infection and colonization of plants by
Fusarium is a function of host susceptibility, time of infection, fungal pathogenicity and meteorological conditions [
12,
13]. The disease occurs during or just after flowering where spores that overwintered in plant debris can germinate and penetrate floral tissue. FHB is driven by weather conditions where wet and warm environments are required for fungal development [
9]. Integrated management using cultivar resistance and FHB-specific fungicide application are the most effective management techniques [
14]. The challenges of fungicide application derive mainly from timeliness of the fungicide application. Prediction tools such as FHB Prediction Center (
http://www.wheatscab.psu.edu) are helping farmers to assess the risk of disease and decide whether or not fungicide application is profitable.
In recent decades, investigators worldwide have carried out extensive studies to develop genetically resistant cultivars. Quantitative trait locus (QTL) such as
Fhb1,
Qfhs.ifa-5A and
QFhs.nau-2DL are widely used as sources of resistance in breeding programs [
12,
15,
16].
Fhb1, for instance, provides resistance against spread of the disease (type II resistance) while
Qfhs.ifa-5A provides resistance to penetration (type I resistance) [
17,
18]. A map-based cloning of
Fhb1 was recently published, opening the possibility of direct cloning and use of that gene [
19]. Furthermore, techniques such as RNA interference (RNAi) for fungicide development and cultivar resistance are promising approaches to controlling FHB [
20]. However, FHB is a quantitative disease, thus an individual QTL approach may not be efficient in controlling this disease, since resistant cultivars have a compilation of major and minor genes that work together to provide resistance.
Plant resistance to diseases will be positively or negatively affected by climate change [
21,
22]. The genetic composition of a variety can provide resistance to disease; however, environmental conditions largely influence whether or not resistance genes will be expressed [
13]. This raises an important question: how responsive are our cultivars to climate change? Are the current sources of resistance to FHB responsive to an increase in temperature? To our knowledge, there are no studies evaluating whether the current sources of FHB resistance will be effective in a warmed environment. In order to answer that question, we conducted over two years an artificially warmed experiment in order to assess the disease response in a large, diverse wheat mapping panel. Our goal was to determine whether or not the QTL used today for FHB resistance would be responsive to disease in an artificially warmed treatment. Specific objectives were: (i) to evaluate phenotypic response to FHB and other disease traits in a large, diverse soft wheat mapping panel under warmed and control conditions; and (ii) to determine whether there were QTL associated with FHB traits under warmed conditions when compared with control conditions based on GWAS analysis.
4. Discussion
Maintaining and/or increasing yield production under climate change is one of the most important challenges of this century. The stress caused by elevation in temperature, changes in rainfall patterns and increases in pests and diseases is predicted to affect crop production significantly [
4,
5,
13]. Field experiments can best assess crop response to the environment due to natural exposure to pest and disease pressure and weather conditions such as rain, temperature and cloud cover. In this study, we simulated increased temperature by artificially warming the rhizosphere and by causing an FHB epidemic, with the goal of assessing changes in plant response under these stresses. We observed that increasing the temperature by 1.8 °C and 2.0 °C in 2016 and 2017 respectively, reduced the length of the winter wheat growing period (
Table 1). Other researchers have reported field studies simulating increases in air temperature that showed an effect on wheat phenology by reducing the pre-anthesis period [
32,
33]. In another study, soil warming conditions have shown a shortening of the total crop growing season in wheat [
7].
Understanding the effects of a warmed environment in crop response is fundamental to achieving sustainable production in the years ahead. Crop models have been used to predict the effects of climate change. These models show a decrease in yield for each degree-Celsius increase in temperature [
4,
11,
34]. Our results showed an increase in yield of 21% in 2016 and a decrease of 6.4% in 2017 under warmed conditions. Increased yield in 2016 could be explained by the experimental design where in 2016 we had only two replications while in 2017 we had four. Since we presented average values for the genotypes in hill plots, the number of replications could affect the final value. Even though crop models predict a reduction in yield in a warmer environment, other researchers have also found an increase in yield. Li et al. [
35], evaluating the effects of soil warming in wheat, observed an increase in yield. Similar results were observed by Tian et al. [
33] in an experiment with air temperature increase tested in winter wheat. Högy et al. [
8] under elevated soil temperature observed no change in grain yield of barley. Variation in yield response to warming agrees with previous studies in our group [
24].
Progress in plant breeding depends, in part, on the heritability of the traits of interest. We were particularly interested in differences in heritability estimates between control and warmed conditions. As expected, based on previous studies [
24], heading date and plant height had high heritability in the control treatment as well as in the warmed treatment. FHB rating, an indication of disease incidence and severity, had a low heritability in the control environment and its heritability under warmed conditions was not significantly different from zero. Heritability of rating in one of our advanced breeding line trials (grown in single row plots in the scab nursery) in 2016 and 2017 was low, averaging 0.32 (data not shown). The other factors that can account for the low heritability of FHB rating in the present study are: (i) hill plots present a different picture to the person rating than do single rows—there is not as much material for the reviewer to look at and rate; and (ii) the very low h
2 rating in the warmed treatment is probably a reflection of the more rapid development of the plant, making it even more difficult to rate at the proper time.
For FDK and DON, repeatability estimates were greater in the warmed treatment than in the control treatment (
Table 5). FDK repeatability estimates were 0.80 and 0.84 under control and warmed conditions, respectively. Similarly, DON repeatability estimates were 0.71 and 0.75 under control and warmed conditions, respectively. High repeatability values indicated reproducibility of the data in the control and warmed treatment. Our results suggest that selection for disease traits such as FDK and DON could be achieved in a warm treatment. Heading date and FHB rating had a moderate correlation of 0.51 in the warming treatment (
Table 3). This result implies that genotypes with a long flowering period had a prolonged period of exposure to the pathogen.
Genome wide association studies (GWAS) provide a tool that breeders can use to investigate a large population of breeding lines and cultivars for association of genomic markers with important agronomic and disease traits. Furthermore, GWAS, by concurrent analysis of genotypic and phenotypic data, allows the detection of QTL across populations with different backgrounds [
36,
37]. Exploring recombination events that occurred years ago in unrelated individuals to identify alleles in linkage disequilibrium with the marker are one of the advantages of GWAS [
38,
39]. The panel used in this study represents soft red winter wheat lines and cultivars from breeding programs distributed across 14 states. A large and diverse panel provides a more realistic assessment of the genetic response to a warmed treatment than would be possible with a single bi-parental population. Several breeding lines and cultivars from the panel have been used as parents in the University of Kentucky breeding program; thus, the results of the GWAS are relevant to possible breeding progress for performance in a warmer treatment in our program.
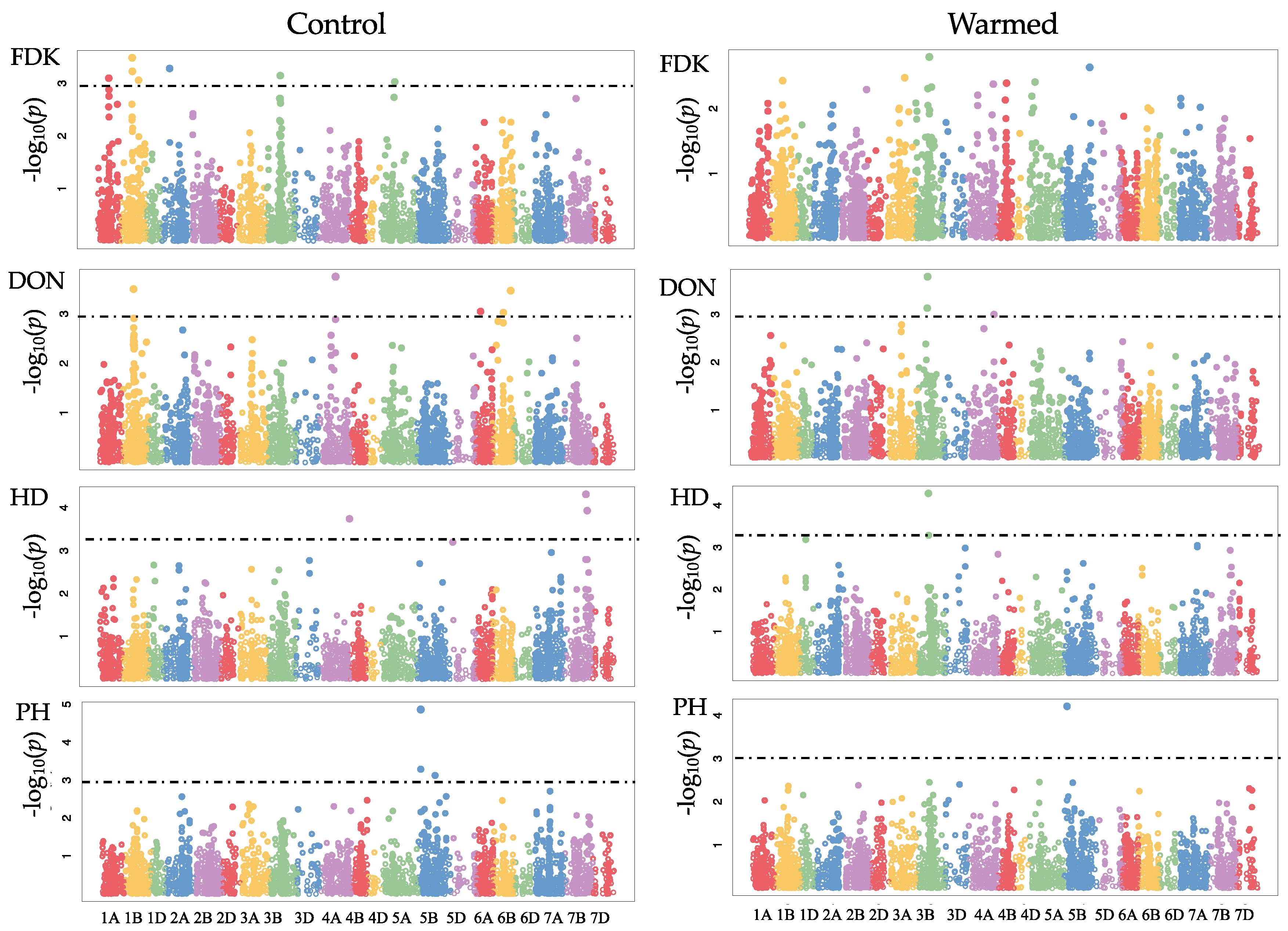
GWAS was carried out for the control and warmed treatments separately. Using a LOD threshold of 3, we detected 19 and 10 SNPs under control and warmed conditions, respectively (
Table 6,
Figure 1). There were significant SNPs for FDK under control conditions with effects ranging from −2.32 to 2.40 on chromosomes 1A, 1B, 2A, 3B, 5A (
Table 6). However, there was no significant SNP for FDK under warmed conditions. In the control treatment, we observed a significant SNP on chromosome 1B with effect of 0.87% on DON levels was founded. In the warmed treatment, the chromosome 3B had two SNPs with effects of 1.13% and 0.94% on DON; neither of these SNPs is associated with
Fhb1. The analysis of HD showed effects ranging from −0.78 to 0.68% under warmed conditions. GWAS revealed potential plant height QTL (not associated with the
Rht loci) with effects from 1.75 to 2.41%. SNP M8584 was detected in the control and warmed treatment, suggesting that this SNP could be used to evaluate populations under warmed conditions.
In complex diseases such as FHB, major and minor genes are involved in conferring levels of resistance [
40,
41]. Plant morphology, resistance to infection and spread of disease as well as environmental factors are critical in determining plant response to pathogens [
17,
18,
42]. The detection of large QTL effects decreases exponentially with the increase in trait complexity [
43]. Thus, for complex traits, with multiple genes involved, an identification of small QTL effects is more likely [
43,
44]. As pointed out by Massman et al. [
44], major QTL in breeding germplasms are under strong selection for multiple years and, thus, fixed in multiple genetic backgrounds. In our study, QTL effects were small, explaining ~−2.5% to +2.6% of the phenotypic variation observed for the traits. If these estimates are accurate, these small effect QTL could be useful in a genomic selection program under warmed conditions.
In addition to the GWAS analysis, we were interested in evaluating the effects of a warmed treatment for disease levels of important wheat QTL (
Table 7).
Fhb1 is widely used in breeding programs to provide resistance to FHB [
19,
45]. In our population of study, under control conditions the presence or absence of this QTL was not statistically significant, which indicates that in these lines the
Fhb1 resistance was not expressed.
Fhb1 increased by 120 and 128%, FDK levels in warmed conditions in absence and presence of the R alleles, respectively. In addition, lines which lacked the resistance alleles at
Fhb1 presented ~69% increase in DON levels under warmed conditions. Traits such as FDK and DON are very important in selecting for FHB resistance since they quantify visually and chemically the level of infection. These traits express phenotypically the effects of resistant QTL such as
Fhb1. Heritability estimates are important in guiding the breeder during selection. The high repeatability values across both environments for FDK and DON are encouraging for selection in a warmed environment (
Table 5).
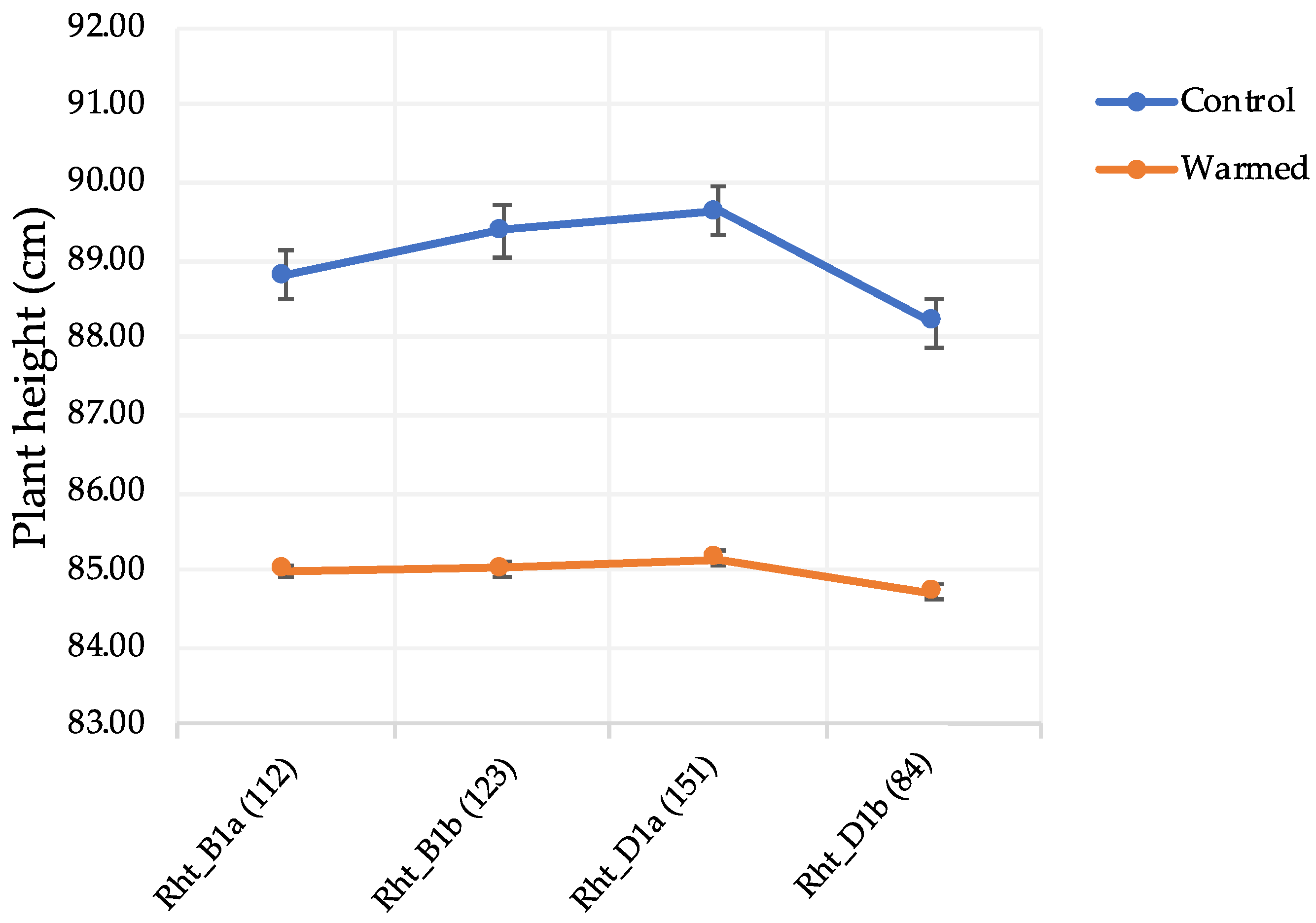
The semi-dwarfing genes (
Rht_B1 and
Rht_D1) had an effect on FDK and DON under warmed conditions. A total of 123 lines had
Rht_B1b (the height reducing allele) in their background. Disease levels for those lines, increased by ~129% and 50% for FDK and DON, respectively, under warmed conditions. Genotypes with
Rht_D1b also showed increased disease levels of ~112% for FDK and ~78% for DON in the warmed treatment (
Table 7). Similar results were observed for tall plants with the wild-type alleles (
Rht_B1a and
Rht_D1a) where the disease increased under warmed conditions (
Table 7). The literature suggests that taller genotypes could maintain a cooler canopy in environments with increased temperatures [
46]. A correlation between increased plant height and reduced FHB is reported in numerous studies [
47,
48,
49]. In one study, the investigators showed that plants with
Rht-B1b and
Rht_D1b significantly decreased resistance to initial infection (Type 1) and
Rht_B1b increased resistance to spread of the fungus [
42]. Under control conditions, in our study, we observed increased disease when the height reducing alleles were present at these QTL. However, the warmed treatment more than doubled the disease rates independent of the allele present (
Table 7). A warmer rhizosphere changed plant morphology by shortening the genotypes which could have favored increased disease levels (
Figure 2). Short plants have higher disease levels due to the microclimate around the spike where high moisture and humidity and close proximity to the inoculum are favorable for increased disease [
50]. Yan et al. [
51] demonstrated this result by studying near-isogenic lines for
Rht genes. His group observed that tall plants were more resistant to infection (type I) than the semi-dwarf phenotypes, however when both phenotypes were at the same height that difference disappeared [
51].
Wheat is cultivated in a wide range of environments due to its vernalization and photoperiod genes. A prerequisite for winter wheat is the accumulation of cold temperature which may be affected by climate change in that warm winters will be more frequent. Vernalization requirements are controlled by
Vrn genes and specific environmental conditions are needed for the activation of these genes [
52,
53]. Zheng et al. [
54] studied the rates of climate change in Australia; they suggested an earlier sowing to escape frost and heat stresses. In addition to earlier sowing, longer season varieties have been proposed as a strategy to adapt to future climate change [
54,
55]. However, longer season varieties in an environment with a high level of disease pressure could increase FHB rates due to increased exposure to the pathogen. Meteorological factors such as wet and warm environments are essential for pathogen development and whether or not such factors will occur in a specific region is difficult to predict [
13]. In this study, we found that plants with
vrn_A1 had 123% more FDK and 60% more DON under warmed conditions (
Table 7). Similar results were observed for
vrn_B1 with vales of 122% for FDK and 62% for DON. In addition, FDK and DON had increases of 120% and 66%, respectively, for
vrn_D3. An analysis of heading date showed that plants under warmed conditions headed earlier than the control (
Table 1). This suggests that plants were exposed earlier to the pathogen increasing the period for disease, which could explain the increased disease levels found in this study.
Photoperiod (
Ppd) genes are another important major gene for the transition from vegetative stage to reproductive stage in plants [
56]. In photoperiod response, the dominant allele confers flowering under short day-length through photoperiod insensitivity [
57]. After vernalization requirements are met, plants with photoperiod insensitivity will shift to the reproductive stage when temperatures increase, while for photoperiod sensitive plants, a long day is also needed [
56]. In the warmed treatment genotypes had an earlier flowering period when compared with the control (
Table 1). An increase in disease levels in the warmed treatment was observed, ranging from 109 to 135% for FDK and from 60 to 65% for DON in the genotypes classified in sensitive and insensitive photoperiod (
Table 7). An earlier heading period favored disease development since the pathogen was present and wet conditions prevailed in the environment. Therefore, a late heading type, characterized by
Ppd sensitivity, probably would not avoid disease in a warmer environment.
The ultimate goal in breeding programs is to develop cultivars with high yield performance and adaptability to multiple environments. Screening for agronomic as well as disease resistance traits is fundamental for the selection of superior genotypes. Future projections of increasing temperature add another degree of complexity to selection. Disease evaluations are complex and highly dependent on environmental conditions. Changes in temperature and rainfall patterns can potentially influence levels of disease, with increased spore production and aggressiveness of the pathogen [
13]. The design of experiments evaluating increases in temperature associated with disease pressure is critical for a better understanding of the genetics behind the phenotypic response. Moreover, the evaluation under warmed conditions of mycotoxin production such as DON is essential for food security.
The Food and Drug Administration (FDA, [
58]) advisory level for DON in finished wheat products such as flour, bran and germ consumed by humans is 1 ppm. Therefore, it is extremely important to develop genotypes that have the ability to fight the infection caused by the pathogen either by morphological characteristics or by resistance QTL [
40,
49]. To assess the variation among cultivars and breeding lines for DON levels, we classified the genotypes in the warmed treatment for best and worst performance for DON levels and ranked them from one to two hundred and thirty-eight. After that, we determined how those genotypes performed in the control treatment and their rank order among the 238 genotypes (
Table 8). Spearman’s rank correlation between warmed and control was 0.56, indicating a moderate correlation between genotype ranks in the two treatments.
The best fifteen performers under warmed conditions had, on average, 2.30 ppm DON with the lowest being 1.70 ppm and the highest 2.78 ppm (
Table 8). Under control conditions, those genotypes had a similar response with 2.48 ppm of DON, on average. The best fifteen in the warmed treatment, had similar performance in the control treatment, with low DON levels. The exception was the genotype 0570A1-2-39-5, which had 135% more DON under control conditions than in the warmed treatment. The best four lines in the TCAP population had levels of DON below 2 ppm in the warmed treatment. Even though the best fifteen lines had DON levels above the FDA advisory level, those lines performed well under warmed conditions and could be indicated for use in breeding programs targeting disease resistance under climate change.
The worst fifteen performing genotypes for DON under warmed conditions and their respective performance under control conditions are presented in
Table 8. The average DON level was 16.74 ppm under warmed conditions and 8.23 ppm under control conditions. DON levels varied from 14.18 to 24.83 ppm under warming and from 3.75 to 14.55 ppm in the control treatment. These results show the dramatic effect of a warmed treatment on genotypes that have low plasticity to an increase in temperature. Genotypes such as MD03W61-11-2 and BECKER had low levels of DON under control conditions; however, under warmed conditions these two lines showed increases of ~290 and 417%, respectively (
Table 8). Similarly, the breeding line VA10W-140 had an increase of ~231% under warmed conditions when compared with its performance under control conditions. Some genotypes had high levels of DON in both treatments. For instance, the MO100535 and 03633A1-69-2-5 had 11.90 and 14.55 ppm under control conditions, respectively. Under warmed conditions, MO100535 had 14.58 ppm and 03633A1-69-2-5 had 19.27 ppm, an increase of ~23 and 32%, respectively. For OH08-149-11 and MD03W665-10-3, DON levels were 10.50 ppm and 10.85 ppm under control conditions, respectively. Under warmed conditions, OH08-149-11 and MD03W665-10-3, had 14.63 ppm and 15.23 ppm an increase of ~39 and 40%, respectively. These genotypes presented high levels of disease and thus would not be indicated for use in breeding programs irrespective of treatment.
As mentioned by Atlin et al. [
59], breeding programs will play a fundamental role in adaptation to climate change, where favorable alleles to environmental stresses will need to be rapidly and constantly incorporated into breeding material to produce genotypes capable of adapting to the environment. Therefore, evaluating our current germplasm for disease resistance under warmed conditions is very important. An artificially warmed treatment can be efficient in screening the genotypes and providing valuable information regarding disease response. In the current study, we demonstrated that an increase in soil temperature of a few degrees resulted in earlier heading. As a consequence, disease levels were higher in the warmed treatment than in the control treatment. Our GWAS analysis identified 19 SNPs in the control treatment and 10 SNPs in the warmed treatment. These SNPs can be useful for selection under warmed conditions. We studied the effect of genotype at important QTL such as
Fhb1,
Rht,
vrn and
Ppd on disease levels in the population. While we did observe some differences between mutant and wild type alleles at certain loci (e.g.,
Rht-D1), the much greater difference in disease levels was observed between the warmed and control treatments, independent of the allelic form present. Evaluation of cultivar/breeding line performance under warmed conditions showed that the superior genotypes under control conditions were often the best performers in the warmed treatment. There were exceptions to this trend, however. For example, KY02C-2215-02, 0570A1-2-39-5, 0513A1-1-3 ranked highly for DON under warming but slipped to ranks 51, 186 and 57 in the control treatment (
Table 8). Falconer [
60] showed that for indirect selection to be superior to direct selection, the genetic correlation must be high and heritability of the trait to be selected must exceed that of the other trait. Using phenotypic correlation as a proxy for genetic correlation and taking the square root of the reliability estimates allows us to estimate Q, the ratio of indirect to direct selection which is expressed as the product of the genetic correlation coefficient and the ratio of the square roots of the heritabilities (e.g., Q = r
g × h
1/h
2; [
60]). In this instance r
p = 0.58, h
1 = 0.84 and h
2 = 0.86 which leads to a product of 0.56, which is less than 1 and clearly does not support indirect selection under control conditions for performance under warming.
These findings, coupled with moderate to high reliability estimates suggest that breeding for disease resistance in a warming treatment should be possible. The active warming method described herein should provide breeders with a tool to pursue this breeding objective.