The Potential of Lr19 and Bdv2 Translocations to Improve Yield and Disease Resistance in the High Rainfall Wheat Zones of Australia
Abstract
:1. Introduction
2. Results
| Year | Site | Average Site Yield | T4 Translocation | TC14 Translocation | ||
|---|---|---|---|---|---|---|
| No. Plots | Genetic Backgrounds | No. Plots | Genetic Backgrounds | |||
| 2000 | Ginninderra a | 4.71 | 192 | 12 | - | - |
| 2000 | Gundibindyal | 6.70 | 120 | 9 | - | - |
| 2000 | Moombooldool | 4.27 | 160 | 11 | - | - |
| 2001 | Condobolin | 1.57 | 80 | 10 | - | - |
| 2001 | Ginninderra a | 6.24 | 80 | 11 | - | - |
| 2001 | Gundibindyal | 4.26 | 96 | 12 | - | - |
| 2001 | Moombooldool | 3.09 | 96 | 11 | - | - |
| 2006 | Griffith a | 4.66 | 23 | 7 | 177 | 13 |
| 2006 | Gundibindyal | n.d. | 6 | 4 | 90 | 11 |
| 2007 | Gundibindyal | 1.55 | 63 | 8 | 225 | 9 |
| 2007 | Yanco a | 3.44 | 29 | 8 | 211 | 9 |
| 2008 | Temora | 1.91 | 63 | 9 | 225 | 9 |
| 2008 | Yanco | 0.74 | 59 | 9 | 213 | 9 |
| 2008 | Yanco a | 3.97 | 61 | 9 | 211 | 9 |
| Total | 14 | 1128 | 1325 | |||
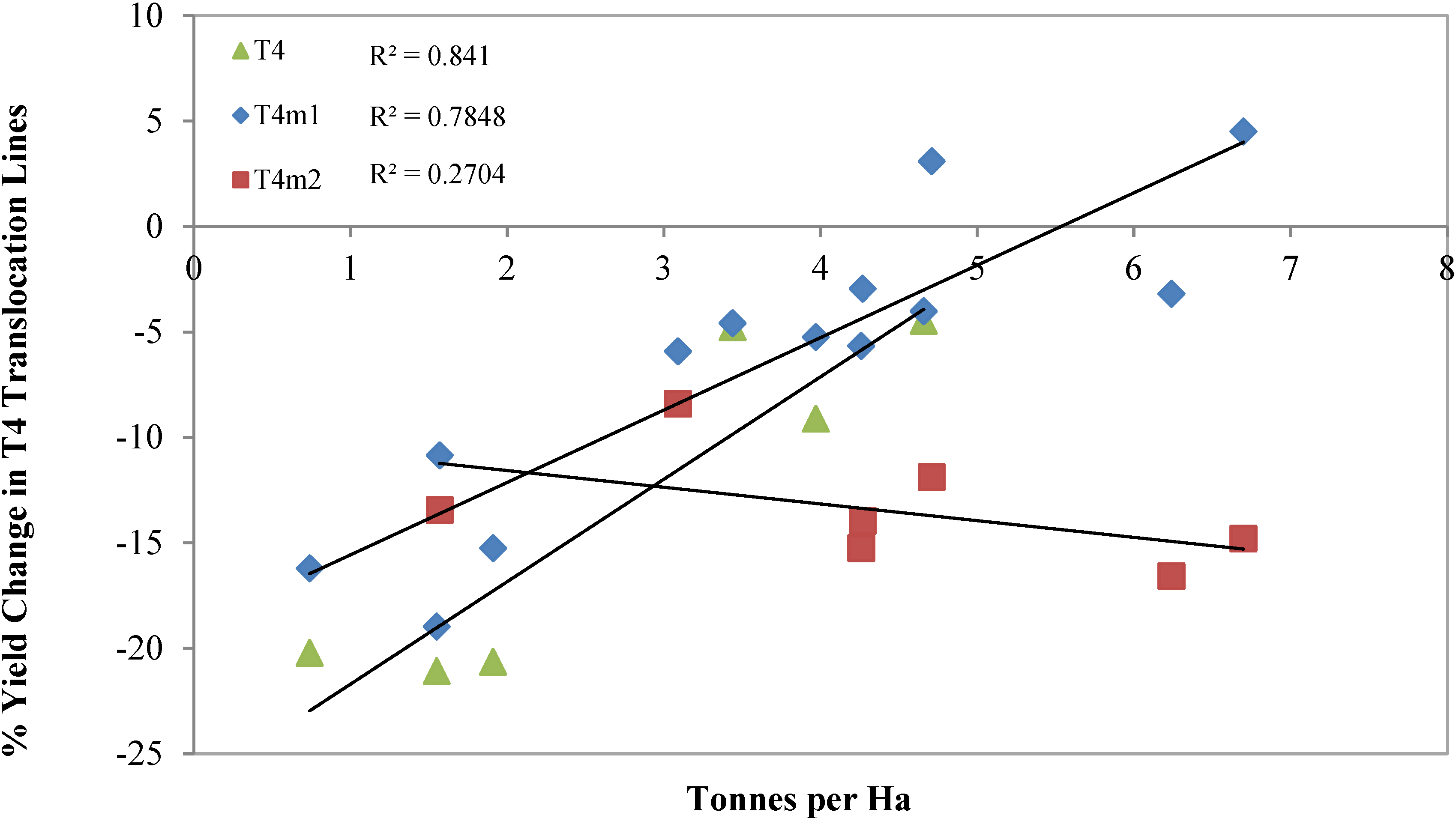
| Environment | Germplasm a | Yield (t/ha) | KGrain·m−2 | TKW (mg) | HI | Total Biomass (t/ha) | Maturity b | Height (cm) |
|---|---|---|---|---|---|---|---|---|
| Ginninderra 2000 c,d | ||||||||
| Recurrent Parents | 4.80 | 61.5 | ||||||
| T4m1 | 5.24 | 58.3 | ||||||
| H2 = 0.81 | T4m2 | 4.46 ** | 58.3 ** | |||||
| Gundibindyal 2000 d | Recurrent Parents | 6.64 | 58.4 | 101.3 | ||||
| T4m1 | 6.93 | 61.2 | 100.7 | |||||
| H2 = 0.89 | T4m2 | 5.32 ** | 58.6 * | 116.1 * | ||||
| Moombooldool 2000 d | Recurrent Parents | 4.35 | 0.429 | 10.48 | 61.5 | 79.8 | ||
| T4m1 | 4.31 | 0.438 | 9.74 | 63.0 | 77.5 *** | |||
| H2 = 0.74 | T4m2 | 3.81 ** | 0.415 * | 9.09 * | 61.6 ns | 87.1 *** | ||
| Condobolin 2001 d | Recurrent Parents | 1.67 | 6017 | 28.1 | 0.357 | 4.74 | 67.2 | 63.4 |
| T4m1 | 1.56 *** | 5786 | 26.6 | 0.341 | 4.50 | 68.7 | 61.5 * | |
| H2 = 0.83 | T4m2 | 1.39 *** | 5420 * | 26.7 | 0.321 *** | 4.42 * | 65.0 ns | 62.8 |
| Ginninderra 2001 c,d | Recurrent Parents | 6.57 | 17981 | 36.9 | 0.430 | 15.28 | 62.3 | 91.0 |
| T4m1 | 6.28 | 18517 | 34.1 *** | 0.406 ** | 15.53 | 61.4 | 90.6 | |
| H2 = 0.86 | T4m2 | 5.49 *** | 17150 * | 32.3 *** | 0.371 *** | 14.75 ns | 59.2 ** | 100.2 *** |
| Gundibindyal 2001 c | Recurrent Parents | 4.57 | 0.393 | 11.64 | 89.2 | |||
| T4m1 | 4.23 ** | 0.380 * | 11.18 | 85.4 *** | ||||
| H2 = 0.68 | T4m2 | 3.82 *** | 0.344 *** | 11.10 * | 90.8 | |||
| Moombooldool 2001 c | Recurrent Parents | 3.33 | 0.384 | 8.81 | 58.8 | 78.9 | ||
| T4m1 | 3.06 ** | 0.375 * | 8.10 | 58.9 | 74.6 ** | |||
| H2 = 0.73 | T4m2 | 2.98 *** | 0.359 *** | 8.33 ** | 57.6 | 83.0 *** | ||
| Gundibindyal 2007 e | T4 negative lines | 1.92 | 5527 | 34.9 | 0.366 | 5.05 | 59.5 | 63.8 |
| T4 | 1.70 *** | 5163 ** | 33.5 | 0.343 * | 4.88 | 63.5 ns | 64.3 | |
| H2 = 0.66 | T4m1 | 1.59 ** | 5152 | 31.4 ** | 0.371 | 4.22 * | 62.0 ns | 62.6 * |
| Temora 2008 e | T4 negative lines | 2.22 | 6006 | 35.5 | ||||
| T4 | 1.85 *** | 5150 | 34.9 | |||||
| H2 = 0.73 | T4m1 | 1.74 *** | 4844 ** | 35.8 * | ||||
| Yanco 2008 c,e | T4 negative lines | 4.00 | 13194 | 30.5 | 69.4 | 82.4 | ||
| T4 | 3.80 | 13626 | 27.9 * | 69.9 | 86.3 ** | |||
| H2 = 0.81 | T4m1 | 3.70 * | 12975 * | 28.7 | 68.9 * | 83.7 | ||
| Yanco 2008e | T4 negative lines | 0.71 | ||||||
| T4 | 0.69 | |||||||
| H2 = 0.47 | T4m1 | 0.56 * |
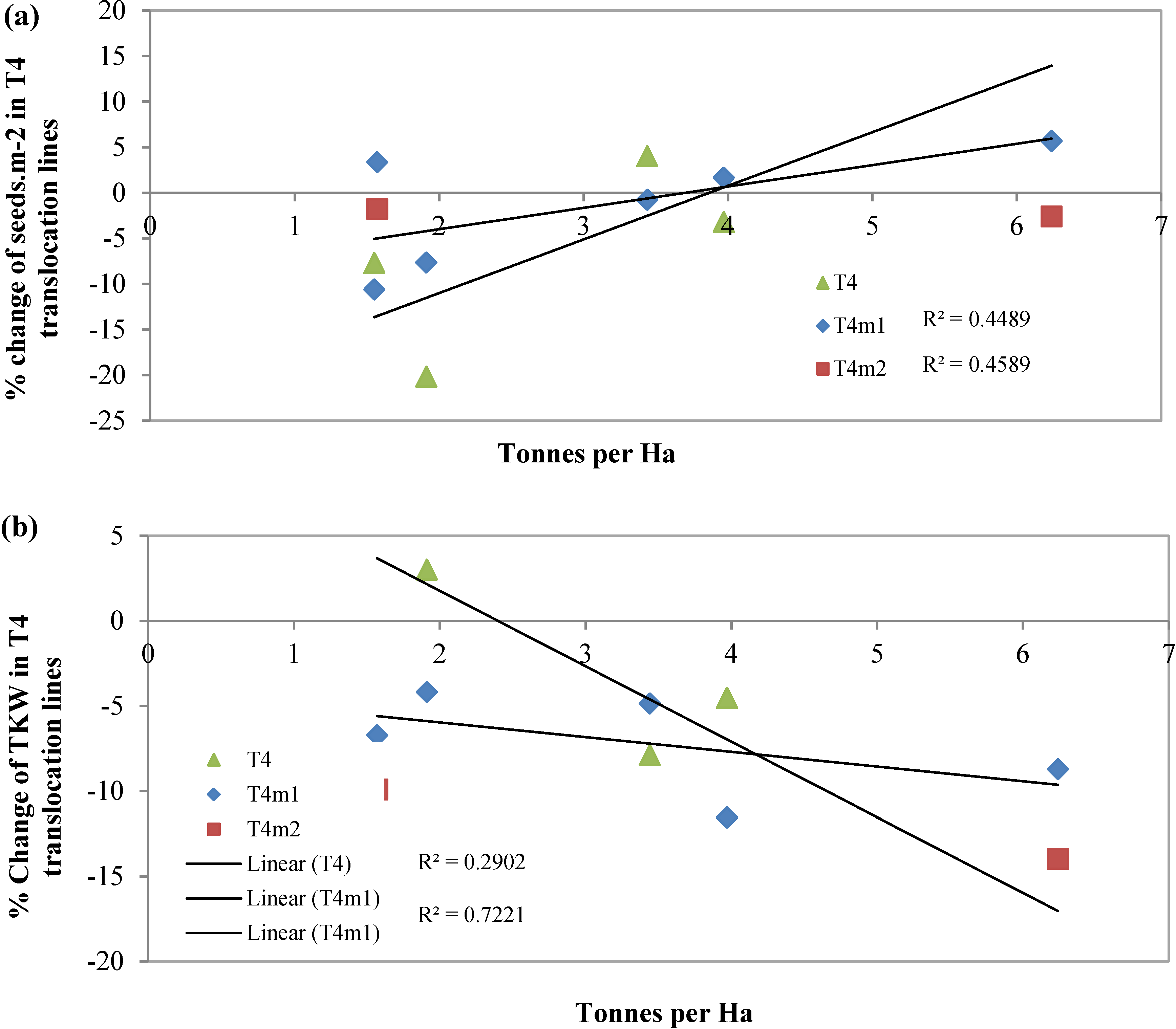
| Site | Germplasm | No. of lines | t·ha−1 | Grains·m−2 | TKW | Maturity a |
|---|---|---|---|---|---|---|
| Temora 2008 | Seri 82/Superseri null | 15 | 2.43 | 7387 | 32.8 | |
| Seri 82/Superseri T4 | 6 | 1.74 *** | 5432 *** | 32.0 | ||
| Condor null | 13 | 2.14 | 6389 | 32.9 | ||
| Condor T4m1 | 10 | 1.93 | 6055 | 31.7 | ||
| Yanco 2008 | Seri 82/Superseri null | 13 | 0.81 | |||
| Seri 82/Superseri T4 | 6 | 0.50 *** | ||||
| Condor null | 10 | 0.58 | ||||
| Condor T4m1 | 10 | 0.54 | ||||
| Yanco 2008 b | Seri 82/Superseri null | 15 | 3.98 | 13484 | 29.53 | 69.13 |
| Seri 82/Superseri T4 | 6 | 3.58 | 13355 | 26.73 *** | 68.67 | |
| Condor null | 10 | 3.77 | 13745 | 27.59 | 69.50 | |
| Condor T4m1 | 10 | 3.38 | 13634 | 24.87 *** | 69.90 |
| Environment | Germplasm a | Grain Yield (t/ha) | Grain Number (no·m−2) | TKW (mg) | Maturity b | Hectolitre Weight | Height (cm) |
|---|---|---|---|---|---|---|---|
| Griffith 2006c | Parents | 4.87 | 57.4 | 87.6 | |||
| Null | 4.55 | 59.1 | 85.7 | ||||
| H2 = 0.72 | TC14 | 4.62 | 55.1 *** | 86.3 | |||
| Gundibindyal 2007 | Parents | 1.43 | 4,012 | 36.2 | 60.0 | 80.0 | 57.3 |
| Null | 1.55 | 4,642 | 34.5 | 60.8 | 79.3 | 60.4 | |
| H2 = 0.59 | TC14 | 1.48 | 4,398 | 34.8 | 59.0 * | 80.0 ** | 60.2 |
| Yanco 2007 c | Parents | 3.82 | 11,362 | 33.9 | 63.7 | 79.7 | 76.3 |
| Null | 3.75 | 11,572 | 32.7 | 63.8 | 80.0 | 75.6 | |
| H2 = 0.60 | TC14 | 3.63 | 11,093 | 33.0 | 61.9 * | 80.1 | 73.0 * |
| Temora 2008 | Parents | 2.34 | 6,757 | 33.6 | 80.4 | ||
| Null | 2.28 | 6,611 | 33.5 | 79.7 | |||
| H2 = 0.46 | TC14 | 2.10 ** | 6,238 | 33.0 | 79.8 | ||
| Yanco 2008 | Parents | 0.69 | |||||
| Null | 0.67 | ||||||
| H2 = 0.49 | TC14 | 0.65 | |||||
| Yanco 2008 c | Parents | 4.19 | 13,494 | 31.4 | 66.4 | 79.6 | 83.3 |
| Null | 4.11 | 14,170 | 29.2 | 68.0 | 78.5 | 81.9 | |
| H2 = 0.71 | TC14 | 3.95 *** | 13,763 * | 29.2 | 65.6 ** | 78.9 | 80.2 |
| Germplasm | No. of Lines a | 2006 Griffith b | 2007 Gundibindyal | 2007 Yanco b | 2008 Yanco b | 2008 Temora | 2008 Yanco | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yield | Maturity | Height | Yield | Maturity | Height | Yield | Maturity | Height | Yield | Maturity | Height | Yield | Yield | ||
| Thelin/4*Camm Null | 12 | - | - | - | 1.81 | 49 | 63 | 3.66 | 63 | 77 | 3.76 | 70 | 77 | 2.68 | 0.62 |
| Thelin/4*Camm TC14 | 12 | - | - | - | 1.70 | 48 | 63 | 3.54 | 62 | 75 | 3.71 | 69 | 78 | 2.39 * | 0.56 |
| Thelin/4*Chara Null | 12 | - | - | - | 1.26 | 47 | 61 | 4.14 | 57 | 75 | 4.52 | 67 | 85 | 2.33 | 0.45 |
| Thelin/4*Chara TC14 | 12 | - | - | - | 1.32 | 44 | 63 | 3.86 | 55 | 70 * | 4.33 | 64 | 82 | 2.28 | 0.46 |
| Thelin/4*Drysdale Null | (15) 11 | 4.87 | 57 | 87 | 2.17 | 48 | 63 | 4.23 | 60 | 75 | 4.25 | 70 | 83 | 1.46 | 0.55 |
| Thelin/4*Drysdale TC14 | (4) 7 | 5.85 | 52 | 96 * | 2.09 | 46 * | 61 | 4.08 | 58 * | 72 | 4.10 | 67 * | 76 | 1.60 | 0.53 |
| Thelin/4*H45 Null | (17) 12 | 4.99 | 67 | 85 | 1.53 | 52 | 67 | 3.85 | 66 | 73 | 4.70 | 70 | 85 | 2.86 | 0.76 |
| Thelin/4*H45 TC14 | (8) 12 | 4.85 | 64 ** | 85 | 1.53 | 52 | 67 | 3.66 | 66 | 73 | 4.33 ** | 70 | 81 ** | 2.66 ** | 0.76 |
| Thelin/4*Janz Null | (5) 12 | 4.65 | 51 | 79 | 1.14 | 48 | 56 | 3.83 | 60 | 73 | 4.23 | 69 | 81 | 2.40 | 0.69 |
| Thelin/4*Janz TC14 | (3) 12 | 4.61 | 49 | 76 | 1.13 | 46 ** | 51 * | 3.85 | 56 * | 72 | 3.87 * | 68 | 78 * | 2.13 | 0.64 |
| Thelin/3*Drysdale//Sunbrook Null | (19) 14 | 4.19 | 56 | 88 | 0.95 | 45 | 53 | 3.71 | 53 | 78 | 4.14 | 60 | 83 | 2.53 | 0.65 |
| Thelin/3*Drysdale//Sunbrook TC14 | (10) 13 | 4.29 | 52 | 86 | 1.15 | 45 | 56 | 3.43 | 51 | 70 * | 4.04 | 60 | 81 | 2.39 | 0.78 |
| Thelin/3*Westonia//Sunbrook Null | (14) 14 | 4.54 | 60 | 84 | 1.75 | 48 | 62 | 3.42 | 59 | 75 | 4.40 | 65 | 79 | 2.21 | 0.75 |
| Thelin/3*Westonia//Sunbrook TC14 | (10) 17 | 4.35 | 54 | 91 | 1.58 | 47 | 61 | 3.74 | 57 | 78 | 4.09 | 59 | 83 | 2.10 | 0.69 |
| Thelin/4*Westonia Null | (10) 12 | 4.69 | 63 | 79 | 1.15 | 54 | 50 | 4.01 | 66 | 70 | 4.37 | 70 | 77 | 2.24 | 0.64 |
| Thelin/4*Westonia TC14 | (9) 12 | 4.86 | 59 | 79 | 1.16 | 52 | 52 | 4.27 | 65 | 74 | 4.38 | 69 | 80 | 1.80 * | 0.54 |
| Thelin/3*H45//Darter Null | (5) | 3.72 | 63 | 91 | - | - | - | - | - | - | - | - | - | - | - |
| Thelin/3*H45//Darter TC14 | (2) | 3.89 | 64 | 95 | - | - | - | - | - | - | - | - | - | - | - |
| Chara/Ohm/Sunbrook Null | (2) | 4.25 | 65 | 74 | - | - | - | - | - | - | - | - | - | - | - |
| Chara/Ohm/Sunbrook TC14 | (2) | 4.94 | 41 ** | 95 | - | - | - | - | - | - | - | - | - | - | - |
3. Discussion
4. Experimental Section
4.1. Development of Germplasm
4.2. Yield Trials
4.3. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.X.; Olsen, O.-A. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345. [Google Scholar] [CrossRef]
- Trethowan, R.M.; Mujeeb-Kazi, A. Novel germplasm resources for improving environmental stress tolerance of hexaploid wheat. Crop Sci. 2008, 48, 1255–1265. [Google Scholar] [CrossRef]
- Knott, D.R. The effect of transfers of alien genes for leaf rust resistance on the agronomic and quality characteristics of wheat. Euphytica 1989, 44, 65–72. [Google Scholar] [CrossRef]
- Sharma, D.; Knott, D.R. The transfer of leaf-rust resistance from agropyron to triticum by irradiation. Can. J. Genet. Cytol. 1966, 8, 137–143. [Google Scholar] [CrossRef]
- Somo, M.; Chao, S.; Acevedo, M.; Zurn, J.; Cai, X.; Marais, F. A genomic comparison of homoeologous recombinants of the Lr19 (T4) translocation in wheat. Crop Sci. 2014, 54, 565–575. [Google Scholar] [CrossRef]
- Singh, R.P.; Huerta-Espino, J.; Rajaram, S.; Crossa, J. Agronomic effects from chromosome translocations 7DL.7AG and 1BL.1RS in spring wheat. Crop Sci. 1998, 38, 27–33. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Calderini, D.F.; Condon, A.G.; Rajaram, S. Physiological basis of yield gains in wheat associated with the Lr19 translocation from agropyron elongatum. Euphytica 2001, 119, 139–144. [Google Scholar] [CrossRef]
- Monneveux, P.; Reynolds, M.P.; Aguilar, J.G.; Singh, R.P.; Weber, W.E. Effects of the 7DL.7AG translocation from lophopyrum elongatum on wheat yield and related morphophysiological traits under different environments. Plant Breed. 2003, 122, 379–384. [Google Scholar] [CrossRef]
- Knott, D.R. Mutation of a gene for yellow pigment linked to Lr19 in wheat. Can. J. Genet. Cytol. 1980, 22, 651–654. [Google Scholar] [CrossRef]
- Banks, P.M.; Larkin, P.J.; Bariana, H.S.; Lagudah, E.S.; Appels, R.; Waterhouse, P.M.; Brettell, R.I.S.; Chen, X.; Xu, H.J.; Xin, Z.Y.; et al. The use of cell culture for subchromosomal introgressions of barley yellow dwarf virus resistance from thinopyrum intermedium to wheat. Genome 1995, 38, 395–405. [Google Scholar] [CrossRef]
- Prins, R.; Groenewald, J.Z.; Marais, G.F.; Snape, J.W.; Koebner, R.M.D. AFLP and STS tagging of Lr19, a gene conferring resistance to leaf rust in wheat. Theor. Appl. Genet. 2001, 103, 618–624. [Google Scholar] [CrossRef]
- Stoutjesdijk, P.; Kammholz, S.J.; Kleven, S.; Matsay, S.; Banks, P.M.; Larkin, P.J. PCR-based molecular marker for the Bdv2 Thinopyrum intermedium source of barley yellow dwarf virus resistance in wheat. Aust. J. Agric. Res. 2001, 52, 1383–1388. [Google Scholar] [CrossRef]
- Ayala-Navarrete, L.; Bariana, H.S.; Singh, R.P.; Gibson, J.M.; Mechanicos, A.A.; Larkin, P.J. Trigenomic chromosomes by recombination of Thinopyrum intermedium and Th. ponticum translocations in wheat. Theor. Appl. Genet. 2007, 116, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Zhang, W.; Dubcovsky, J. Association between allelic variation at the phytoene synthase 1 gene and yellow pigment content in the wheat grain. Theor. Appl. Genet. 2008, 116, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Prins, R.; Marais, G.F.; Marais, A.S.; Janse, B.J.H.; Pretorius, Z.A. A physical map of the thinopyrum-derived Lr19 translocation. Genome 1996, 39, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Prins, R.; Marais, G.F.; Pretorius, Z.A.; Janse, B.J.H.; Marais, A.S. A study of modified forms of the Lr19 translocation of common wheat. Theor. Appl. Genet. 1997, 95, 424–430. [Google Scholar] [CrossRef]
- Ayala-Navarrete, L.I.; Mechanicos, A.A.; Gibson, J.M.; Singh, D.; Bariana, H.S.; Fletcher, J.; Shorter, S.; Larkin, P. The pontin series of recombinant alien translocations in bread wheat: Single translocations integrating combinations of Bdv2, Lr19 and Sr25 disease-resistance genes from Thinopyrum intermedium and Th. ponticum. Theor. Appl. Genet. 2013, 126, 2467–2475. [Google Scholar] [CrossRef] [PubMed]
- Slafer, G.A. Differences in phasic development rate amongst wheat cultivars independent of responses to photoperiod and vernalization. A viewpoint of the intrinsic earliness hypothesis. J. Agric. Sci. 1996, 126, 403–419. [Google Scholar] [CrossRef]
- Huerta-Espino, J.; Singh, R.P. First report of virulence to wheat with leaf rust resistance gene Lr19 in Mexico. Plant Dis. 1994, 78, 640. [Google Scholar] [CrossRef]
- Bhardwaj, S.C.; Prashar, M.; Kumar, S.; Jain, S.K.; Datta, D. Lr19 resistance in wheat becomes susceptible to Puccinia triticina in India. Plant Dis. 2005, 89, 1360–1360. [Google Scholar] [CrossRef]
- Murray, G.M.; Brennan, J.P. The Current and Potential Costs from Diseases of Wheat in Australia; Grains Research and Development Corporation: Kingston, Australia, 2009. [Google Scholar]
- Cullis, B.R.; Thomson, F.M.; Fisher, J.A.; Gilmour, A.R.; Thompson, R. The analysis of the NSW wheat variety database. 2. Variance component estimation. Theor. Appl. Genet. 1996, 92, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, A.R.; Gogel, B.J.; Cullis, B.R.; Thompson, R. Asreml User Guide Release 3.0; VSN International Ltd.: Hemel Hempstead, UK, 2001. [Google Scholar]
- Cullis, B.; Gogel, B.; Verbyla, A.; Thompson, R. Spatial analysis of multi-environment early generation variety trials. Biometrics 1998, 54, 1–18. [Google Scholar] [CrossRef]
- Smith, A.; Cullis, B.; Thompson, R. Analyzing variety by environment data using multiplicative mixed models and adjustments for spatial field trend. Biometrics 2001, 57, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martínez, C.T. Estimating and interpreting heritability for plant breeding: An update. In Plant Breeding Reviews; John Wiley & Sons, Inc.: Raleigh, CO, USA, 2003; pp. 9–112. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosewarne, G.; Bonnett, D.; Rebetzke, G.; Lonergan, P.; Larkin, P.J. The Potential of Lr19 and Bdv2 Translocations to Improve Yield and Disease Resistance in the High Rainfall Wheat Zones of Australia. Agronomy 2015, 5, 55-70. https://doi.org/10.3390/agronomy5010055
Rosewarne G, Bonnett D, Rebetzke G, Lonergan P, Larkin PJ. The Potential of Lr19 and Bdv2 Translocations to Improve Yield and Disease Resistance in the High Rainfall Wheat Zones of Australia. Agronomy. 2015; 5(1):55-70. https://doi.org/10.3390/agronomy5010055
Chicago/Turabian StyleRosewarne, Garry, David Bonnett, Greg Rebetzke, Paul Lonergan, and Philip J. Larkin. 2015. "The Potential of Lr19 and Bdv2 Translocations to Improve Yield and Disease Resistance in the High Rainfall Wheat Zones of Australia" Agronomy 5, no. 1: 55-70. https://doi.org/10.3390/agronomy5010055






