1. Introduction
Nitrogen deficiency is a widespread problem in many soils requiring the use of nitrogen fertilizers to increase crop productivity. Unfortunately, the widespread use of relatively expensive chemical fertilizers is beyond the reach of most farmers in developing countries. However, a less expensive nitrogen source is available to farmers in the form of the fixation of atmospheric nitrogen by root nodule bacteria (rhizobia) in food and forage legumes [
1]. The energy required to drive atmospheric nitrogen reduction in nodules is produced from sucrose that originates from photosynthesis. Sucrose is translocated from the leaf to the nodule. In return, the symbiotic nitrogen fixation activities of the nodules provide the plant with organic nitrogen. Symbiotic nitrogen assimilation not only spares existing soil nitrogen supplies from depletion, but it acts as a mechanism of green soil fertilization that improves the organic content of soil and so makes a valuable contribution to sustainable agriculture [
2]. Improving our current understanding of the factors that limit the effective establishment and duration of symbiosis is key to maximizing legume production by resource-poor farmers, especially in soils with low nitrogen. Some legumes are more achieve symbiotic nitrogen assimilation more effectively than others. For example, grain legumes, such as peanuts, cowpeas, soybeans, and also fava beans have high rates of nitrogen fixation, particularly under conditions of low soil nitrogen. The presence of nitrogen in the spoil inhibits nodule nitrogen fixation. The tropical legume, soybean is often considered to be the most important grain legume in terms of its economic value. It is a vital source of vegetable protein for food and animal feed world-wide. Soybean nodules are classified as having determinate growth and structure. Symbiotic nitrogen assimilation in soybean nodules produces ureides, which are exported to the plant tissues. The ureides are converted into amino acids, which are used for the synthesis of a wide range of essential molecules, such as nucleotides and proteins.
The determinate nature of soybean nodules means that they lose meristematic activity soon after formation and, hence, nodule growth is based largely on cell expansion that results in the formation of the mature nodule. Soybean nodules are further classified into crown nodules and lateral nodules, depending on where they develop and are positioned on the root system. Crown nodules develop from the stem or tap root whereas lateral nodules develop from the lateral roots, as illustrated in
Figure 1A. In general, nodules have a defined and limited functional lifespan and they tend to senesce early relative to other major plant organs, particularly the leaves. Nodule senescence is associated with a decrease in symbiotic nitrogen fixation, a degradation of leghemoglobin, and the breakdown of symbiosis (
Figure 1B) [
3]. The lifespan of nodules on fast growing annual legumes, such as soybean, is about 10–12 weeks from the point of initiation to senescence [
4]. New nodules can be produced on the lateral roots as the crown nodule senesce leading to a heterogeneous population in terms of age, the oldest nodules on the soybean plant being the crown nodules that form on the main root.
Figure 1.
(A) Crown and lateral nodules on soybean (cultivar Prima 2000). Crown nodules develop early on the tap root (photo taken by E Cruywagen, University of Pretoria); (B) Sections of crown nodules at 4 weeks have a pink interior because of the presence of leghemoglobin associated with active nitrogen-fixation. At 14 weeks the interior becomes green as nodule senescence progresses from the center outwards. By 18 weeks the symbiosis has ended and the interior of the dying nodules becomes brown because of leghemoglobin degradation.
Figure 1.
(A) Crown and lateral nodules on soybean (cultivar Prima 2000). Crown nodules develop early on the tap root (photo taken by E Cruywagen, University of Pretoria); (B) Sections of crown nodules at 4 weeks have a pink interior because of the presence of leghemoglobin associated with active nitrogen-fixation. At 14 weeks the interior becomes green as nodule senescence progresses from the center outwards. By 18 weeks the symbiosis has ended and the interior of the dying nodules becomes brown because of leghemoglobin degradation.
In contrast to the initiation of symbiosis and the first stages of nodule initiation, which have been intensively studied in recent years, very few studies have focused on the process of nodule senescence, which has been hardly been investigated in comparison. Although symptoms and progression of the processes of nodule death have been well described, the nature of the factors and mechanisms that control nodule senescence are largely unknown. Moreover, nodules can suffer premature senescence that is induced in response to abiotic stress factors, such as drought, and also by nitrate fertilization. The stress-induced changes that adversely affect nodule stability and performance are not well studied or understood. Premature nodule senescence shares some common characteristics with the developmental senescence process [
5], starting at the center of the nodule and extending to the periphery [
4]. Senescence in nodules as in other plant organs can be defined as a programmed degeneration process that is the final step of development leading to death [
6]. The senescence process is characterized by the degradation of many metabolic and structural proteins, the nitrogen within the nodules being remobilized and translocated to the flowers to drive reproductive growth [
7].
The induction of cysteine proteases during nodule senescence, which was first reported for peas, is a key process in the later stages of nodule development leading to proteolysis [
8]. As in other organs, the induction of cysteine proteases is an excellent marker for nodule senescence [
8]. The regulated turnover of proteins is important for cell functions and is an intrinsic part of the development process. It underpins cellular homeostasis and is required for the induction of programmed cell death (PCD) at the end of senescence [
9]. All proteins are subject to turnover, involving degradation by the proteolytic systems of the cell including the proteosome and the suite of proteolytic enzymes. Proteins turn over at different rates according to their functions and sensitivity to degradation, which is enhanced during stress or when the nitrogen or carbon from the component amino acids is needed for other processes [
10]. Cysteine proteases contain a catalytic sulfhydryl group at the active center involving a cysteine and a histidine residue. The majority of plant cysteine proteases belong to either the papain (C1) or legumain (caspases) (C13) families with legumains containing similar protein folds to caspases, which belong to the C14 family.
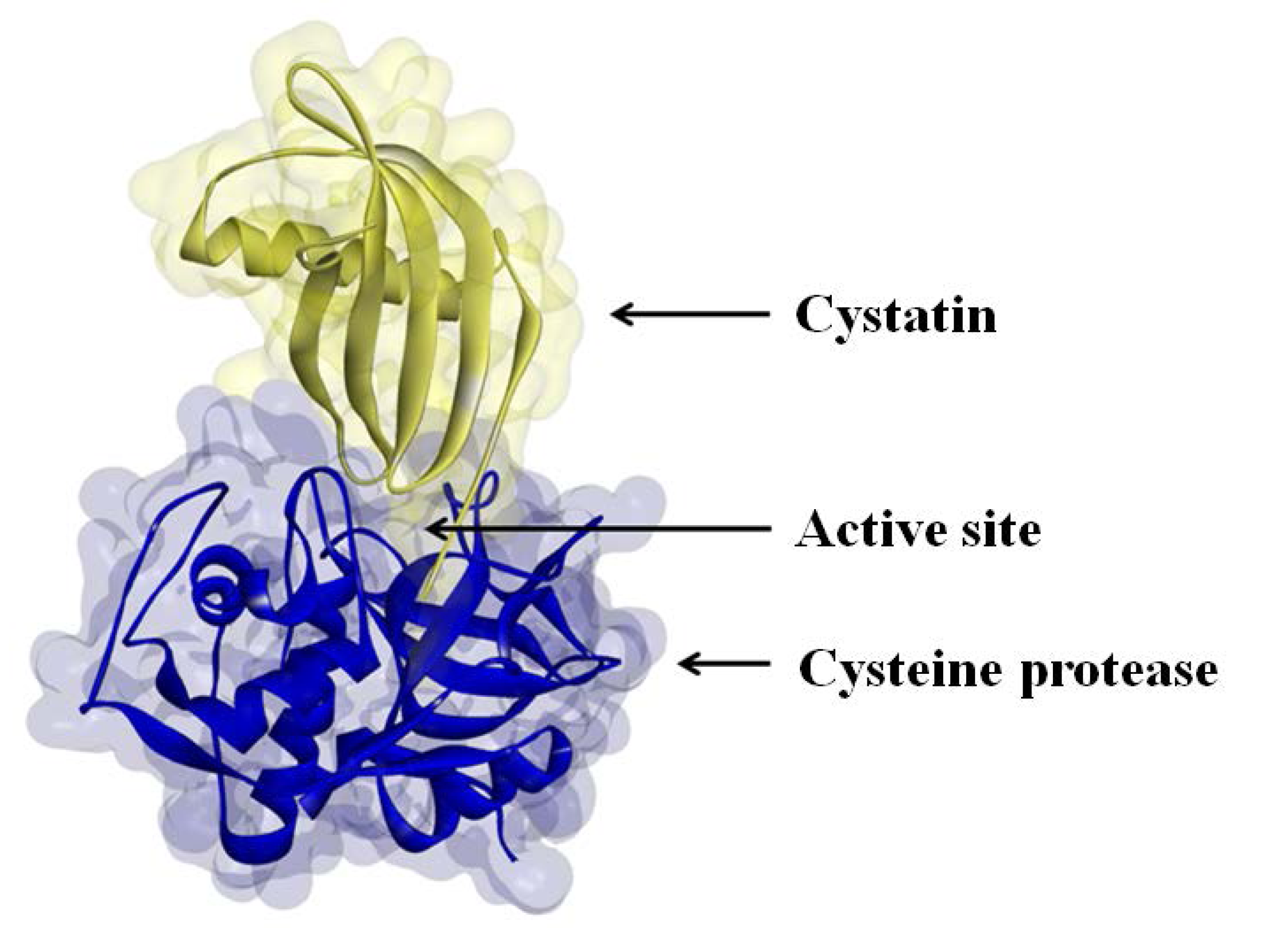
The activities of many proteases are regulated by protease inhibitors, which are generally small proteins that inactivate their protease substrates in a specific manner by forming reversible complexes (
Figure 2) [
11]. In the soybean nodule, the zone of high cysteine protease expression increases in size as the nodule develops and moves from the outer periphery toward the center of the organ [
3]. Although the activities of cysteine proteases have been demonstrated during nodules senescence [
12], little information is available about the genes that encode the cysteine protease forms that are expressed during early and late soybean nodule development. Moreover, to our knowledge, no detailed information exists in the literature concerning the expression of cysteine protease inhibitors (cystatins) during nodule development or in natural or stress-induced senescence.
Figure 2.
Cystatin binding to the active site of a cysteine protease.
Figure 2.
Cystatin binding to the active site of a cysteine protease.
Various biochemical and molecular tools have been used to characterize the cysteine proteases that are involved in nodule senescence. This includes the identification of activities and some analysis of gene expression profiles. In the following discussion, we summarize recent achievements in the characterization of the soybean nodule cysteine protease-cystatin system in relation to agronomic traits. As well as summarizing the relevant literature, we have included information from our own studies using different “omics” tools to characterize the cysteine proteases and cystatins that are specifically expressed at different stages of nodule development.
3. Cysteine Protease and Cystatin Activity in Soybean Nodules
Cysteine proteases and cystatins have been characterized in soybean crown nodules using both biochemical and transcript profiling approaches. Total nodule protease activities were measured in these studies using mildly denaturing gelatin-containing SDS-PAGE gel electrophoresis, in which proteins are not fully denatured and retain activity following separation on the gels [
38].
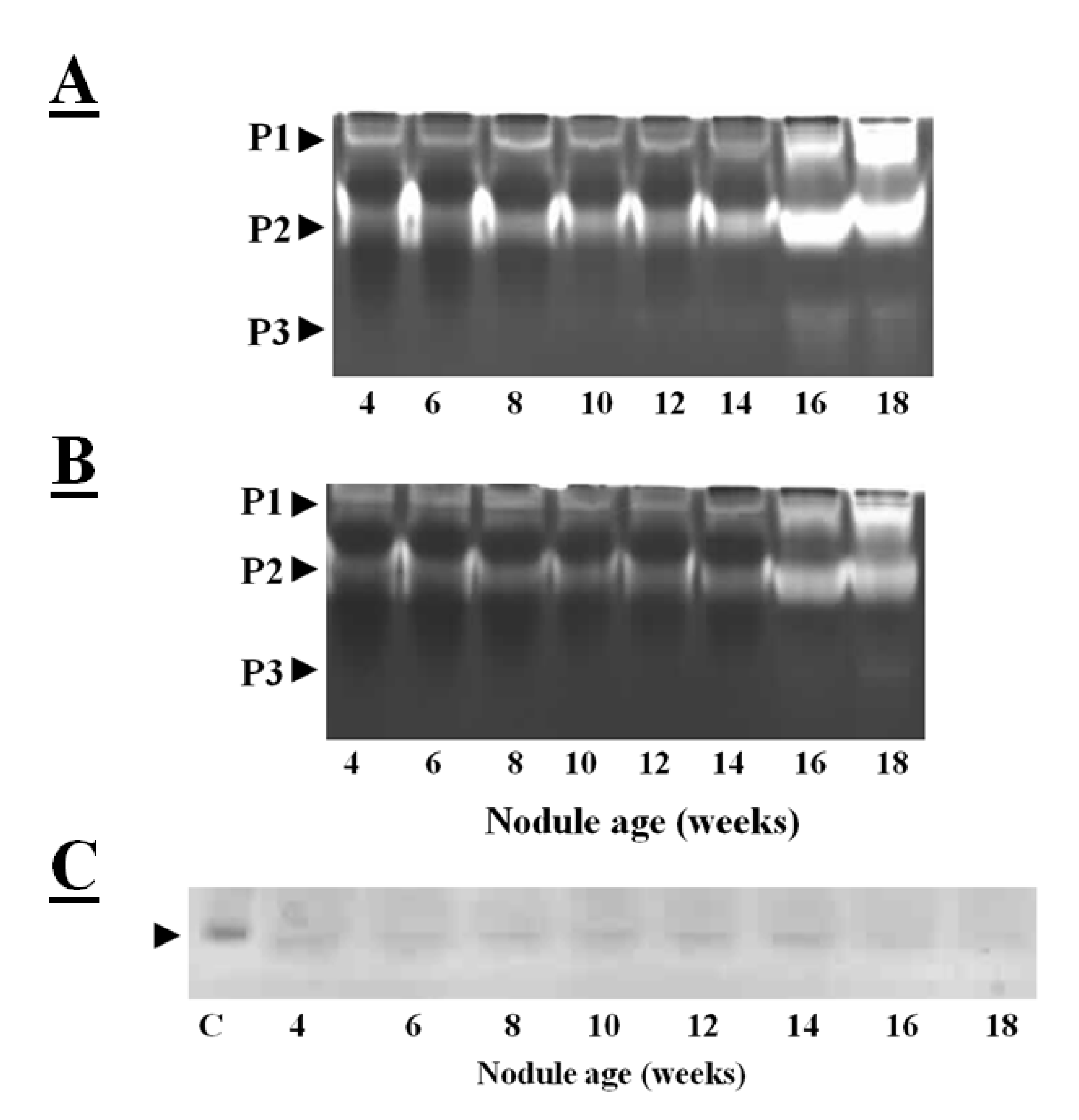
Figure 3A illustrates the cysteine protease activities of soybean crown nodules as observed on gelatin-containing SDS-PAGE gels, where the unstained area has escaped gelatin degradation by proteases. The protease activities of soybean crown nodules of different ages measured in this way show almost identical patterns of total protease activity at least in samples up to an age of 10 weeks (
Figure 3A; protease activity P1 and P2). In contrast, the older, senescent crown nodules showed an additional protease activity (P3) band with a higher protease activity in the P1 and P2 bands demonstrated by a more extensive clearance of the Coomassie blue-stained gel. When E-64, a potent inhibitor of papain-like cysteine proteases [
39] was added to the gels less band clearance was observed in the senescent nodules. This observation would suggest that both E-64-sensitive proteases, most likely papain-like cysteine proteases, as well as cysteine proteases that are not sensitive to E-64 are expressed during soybean nodule senescence (
Figure 3B).
The reverse zymogram technique can be used to detect the presence of cystatins in nodule extracts. This technique is based the ability of nodule cystatins to prevent the papain-catalyzed degradation of gelatin. After the electrophoretic separation of nodule proteins, the gelatin-containing gels were incubated with papain in order to degrade gelatin and nodule proteins. The gels are then stained with Coomassie blue and areas where nodule cystatins block papain, the gelatin remains stained [
38]. Application of this technique revealed that cystatins are expressed in soybean nodules throughout development, except at the senescent stage (16–18 weeks old) where protease activities are high (
Figure 3C). Down-regulation of transcription of cystatins and up-regulation and activation of cysteine proteases might have resulted in higher cysteine protease activity.
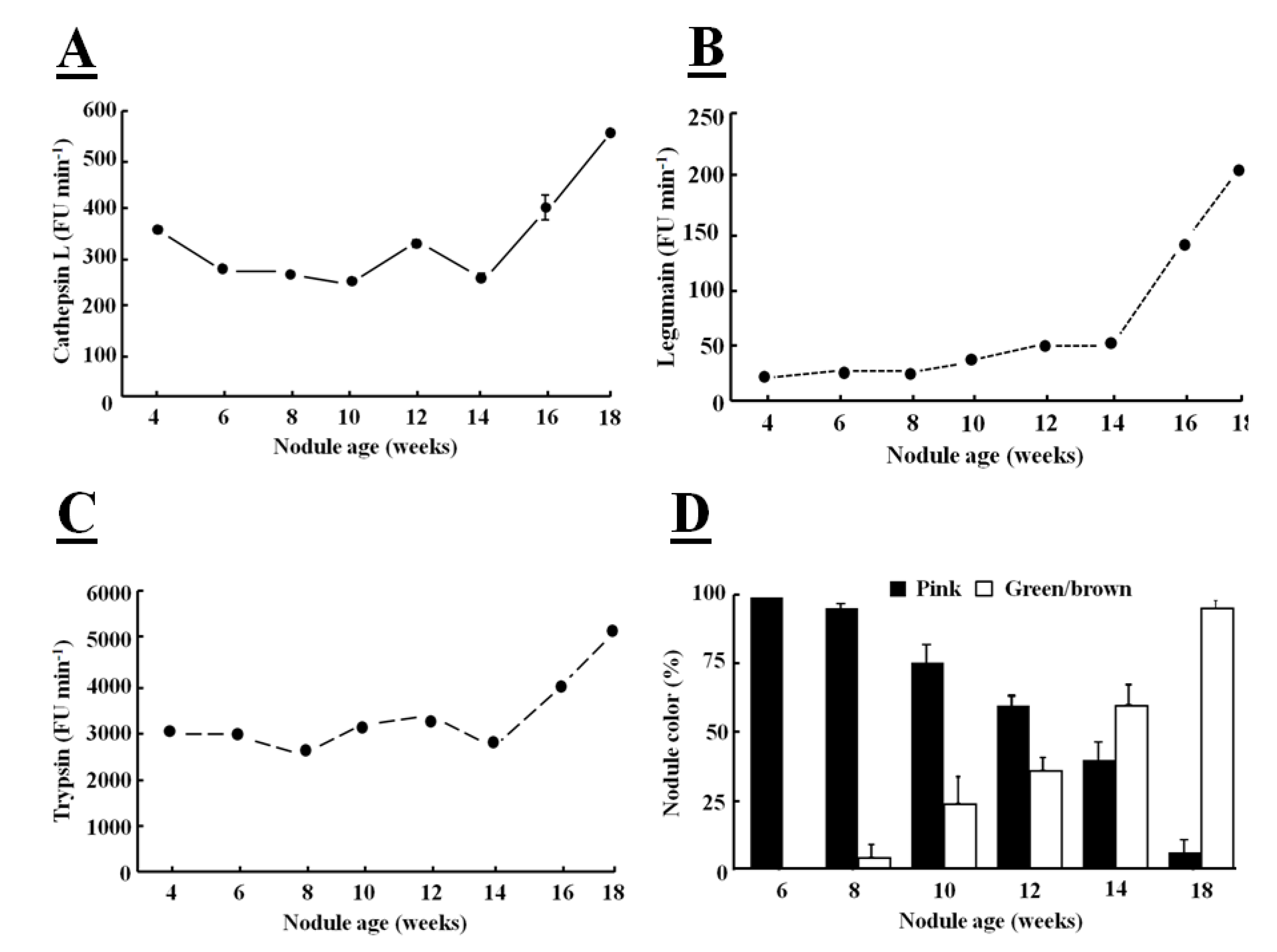
Fluorogenic substrates can be used to detect Phe-Arg-MCA cathepsin L-like, Z-Ala-Ala-Phe-MCA legumain-like and Z-Arg-MCA trypsin-like protease activities [
39]. When these fluorogenic substrates are hydrolyzed by a protease, a bound α-amino 4-methylcoumarin (MCA) derivative is released and this fluoresces in the unbound form. The hydrolysis of all these substrates was greatly increased in senescent nodules confirming that protease activities are increased at the later stages of development (
Figure 4).
The major targets of proteases in root nodules are considered to be cytosolic proteins, such as leghemoglobin, which is the most abundant nodule protein, as part of symbiosome degradation [
40]. The increases in protease activity, particularly in legumain activity, observed during nodule senescence occurred at the same time as leghemoglobin degradation (
Figure 4D), as indicated by the green coloration of the nodules. The color change also indicates that leghemoglobin degrades from the center of the nodule, where the tissues change from pink to green/brown (
Figure 1B). Nodule greening is due to biliverdin formation [
41]. This color change has frequently been used as a visible marker of nodule senescence [
17]. The primary role of leghemoglobin is the transport of oxygen, the high sensitivity of the nitrogenase enzyme in the
Rhizobium bacteria to oxygen-dependent inactivation requiring tight control of tissue oxygen levels. The decrease in nitrogen fixation is associated with a decline in the ability to transport oxygen [
42] as leghemoglobin degradation is initiated (
Figure 5B).
Figure 3.
Total protease activities measured in soybean crown nodules (cultivar Prima 2000). Protease activities were visualized on a mildly denaturing gelatin-containing SDS-polyacrylamide gel over a period of 18 weeks without (
A) and with the addition of E-64 (added to the sample before electrophoresis to a final concentration 10 µM). This detects the presence of both E-64 sensitive proteases (papain like cysteine proteases) as well as proteases from other classes not sensitive to E-64; (
B) Reverse zymogram to detect cystatin activity against papain action (
C); Different protease activities (P1, P2, and P3) are shown by clearance of stained gel. Gel electrophoresis was carried out with A Bio-Rad (UK) electrophoresis system on a 15% (w/v) sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) according to the method of Laemmli [
43] containing 1% (w/v) of gelatin with 50 µg of protein per sample loaded. Protease re-naturing, gel treatment with papain (2mg/100mL of buffer, Sigma, USA) for reverse zymography for protease inhibitor detection and all gel staining procedures were carried out according to the method described by Michaud [
38]. P represents detected protease activity and C oryzacystatin-I (1 µg) used as a control.
Figure 3.
Total protease activities measured in soybean crown nodules (cultivar Prima 2000). Protease activities were visualized on a mildly denaturing gelatin-containing SDS-polyacrylamide gel over a period of 18 weeks without (
A) and with the addition of E-64 (added to the sample before electrophoresis to a final concentration 10 µM). This detects the presence of both E-64 sensitive proteases (papain like cysteine proteases) as well as proteases from other classes not sensitive to E-64; (
B) Reverse zymogram to detect cystatin activity against papain action (
C); Different protease activities (P1, P2, and P3) are shown by clearance of stained gel. Gel electrophoresis was carried out with A Bio-Rad (UK) electrophoresis system on a 15% (w/v) sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) according to the method of Laemmli [
43] containing 1% (w/v) of gelatin with 50 µg of protein per sample loaded. Protease re-naturing, gel treatment with papain (2mg/100mL of buffer, Sigma, USA) for reverse zymography for protease inhibitor detection and all gel staining procedures were carried out according to the method described by Michaud [
38]. P represents detected protease activity and C oryzacystatin-I (1 µg) used as a control.
![Agronomy 03 00550 g003]()
Figure 4.
Determination of cathepsin Ll-like (
A), legumain-like (
B), trypsin-like (
C) protease activities in soybean crown nodules (cultivar Prima 2000) and crown nodules color (
D) over a period of 18 weeks. Cathepsin L-like activity with substrate Z-Phe-Arg-MCA was measured in 50 mM sodium phosphate buffer, pH 6.0, containing 5 mM
l-cysteine, legumain-like cysteine protease activity with substrate Z-Ala-Ala-Asn-MCA measured in buffer containing 39.5 mM citric acid, 121 mM Na
2HPO
4, 1 mM DTT, 1 mM EDTA, and 0.01% CHAPS at pH 7.0 and trypsin-like activity with substrate Z-Arg-MCA in 100 mM Tris-HCl buffer at pH 8.0. Hydrolysis of the substrate was monitored using a hydrolysis progress curve [
44]. Hydrolysis was monitored at 25 °C with a spectro-fluorometer (BMG FluoStar Galaxy) with excitation and emission at 340±10 nm and 450±10 nm, respectively. The reaction was measured over a period of 20 min. Reaction rates represented by the slope of the curve were measured as Fluorescence Units (FU/min). All reactions were performed in triplicate. For nodule color pink color indicates active leghemoglobin, greenish/brown color inactive leghemoglobin. Color values shown represent the mean of four individual experiments ± SE and protease activity data represent the mean ± SE of three individual activity measurements with 2 µg of nodule protein extract added to each assay.
Figure 4.
Determination of cathepsin Ll-like (
A), legumain-like (
B), trypsin-like (
C) protease activities in soybean crown nodules (cultivar Prima 2000) and crown nodules color (
D) over a period of 18 weeks. Cathepsin L-like activity with substrate Z-Phe-Arg-MCA was measured in 50 mM sodium phosphate buffer, pH 6.0, containing 5 mM
l-cysteine, legumain-like cysteine protease activity with substrate Z-Ala-Ala-Asn-MCA measured in buffer containing 39.5 mM citric acid, 121 mM Na
2HPO
4, 1 mM DTT, 1 mM EDTA, and 0.01% CHAPS at pH 7.0 and trypsin-like activity with substrate Z-Arg-MCA in 100 mM Tris-HCl buffer at pH 8.0. Hydrolysis of the substrate was monitored using a hydrolysis progress curve [
44]. Hydrolysis was monitored at 25 °C with a spectro-fluorometer (BMG FluoStar Galaxy) with excitation and emission at 340±10 nm and 450±10 nm, respectively. The reaction was measured over a period of 20 min. Reaction rates represented by the slope of the curve were measured as Fluorescence Units (FU/min). All reactions were performed in triplicate. For nodule color pink color indicates active leghemoglobin, greenish/brown color inactive leghemoglobin. Color values shown represent the mean of four individual experiments ± SE and protease activity data represent the mean ± SE of three individual activity measurements with 2 µg of nodule protein extract added to each assay.
![Agronomy 03 00550 g004]()
Increased protease activities in senescent nodules are associated with both a decrease in nodule protein content and in nitrogenase activity (
Figure 5C,D). The nitrogen-fixing capacity of nodules peaks early in the soybean nodule development (
Figure 5D). Despite some drawbacks, the acetylene reduction assay method for the detection of nitrogenase activity is widely used because of its high sensitivity and simplicity. Nodule weight and number are also frequently used as markers of nitrogen fixation capacity [
45]. Although the fresh weight of soybean nodules increased with time, the number of crown nodules was fixed early in development was constant thereafter (
Figure 5A,B). However, the crown nodules increase in weight during development with the larger nodules fixing more nitrogen than the smaller nodules [
46].
Figure 5.
Production of nodule number (
A), biomass (
B), total protein content (
C) and nitrogenase activity of soybean crown nodules (
D) (cultivar Prima 2000) measured as µmol acetylene reducedmin
−1 g FW
−1 over a period of 16 weeks. For nodule formation soybean seeds (Glycine max L. Merr.; cultivar Prima 2000) were inoculated with
Bradyrhizobium japonicum (strain WB 74-1) powder at 109 CFU g
−1 (Soygro bio-fertilizer Limited, South Africa) and grown in pots [17.5 cm × 20 cm diameter (top) and 13.1 cm] in fine-grade vermiculite (Mandoval PC, South Africa) in an environmentally controlled green-house with a 13 h photoperiod at 25 °C/16 °C day/night temperature, 600 mmol m
−2 s
−1 photosynthetically active radiations (PAR) and 60% relative humidity and watered twice a week with de-ionized water and three-times-a-week with a nitrogen-free Hoagland solution. Protein content of nodule extracts was determined using a commercial protein determination kit (Bio-Rad, UK). Nitrogenase activity of nodules was determined with a gas chromatograph (Varian 3900; Varian Inc., USA) according to the method described by Turner and Gibson [
47]. Data represent the mean ± SD of fresh weight and nodule number derived from three individual plants. Data represent the mean ± SD of activity derived from nodules of three individual plants.
Figure 5.
Production of nodule number (
A), biomass (
B), total protein content (
C) and nitrogenase activity of soybean crown nodules (
D) (cultivar Prima 2000) measured as µmol acetylene reducedmin
−1 g FW
−1 over a period of 16 weeks. For nodule formation soybean seeds (Glycine max L. Merr.; cultivar Prima 2000) were inoculated with
Bradyrhizobium japonicum (strain WB 74-1) powder at 109 CFU g
−1 (Soygro bio-fertilizer Limited, South Africa) and grown in pots [17.5 cm × 20 cm diameter (top) and 13.1 cm] in fine-grade vermiculite (Mandoval PC, South Africa) in an environmentally controlled green-house with a 13 h photoperiod at 25 °C/16 °C day/night temperature, 600 mmol m
−2 s
−1 photosynthetically active radiations (PAR) and 60% relative humidity and watered twice a week with de-ionized water and three-times-a-week with a nitrogen-free Hoagland solution. Protein content of nodule extracts was determined using a commercial protein determination kit (Bio-Rad, UK). Nitrogenase activity of nodules was determined with a gas chromatograph (Varian 3900; Varian Inc., USA) according to the method described by Turner and Gibson [
47]. Data represent the mean ± SD of fresh weight and nodule number derived from three individual plants. Data represent the mean ± SD of activity derived from nodules of three individual plants.
![Agronomy 03 00550 g005]()
4. Characterization of the Cysteine Protease and Cystatin Transcriptome
Transcriptome profiling has been used to characterise the genes involved in the senescence of indeterminate nodules of
Medicago truncatula [
48]. Four cysteine proteases genes were highly expressed in nodule senescence. These genes were highly homologous to one of the most frequently used markers of leaf senescence, the cysteine protease encoded by the gene called SENESCENCE ASSOCIATED GENE 12 (
SAG12) [
19]. An early transcriptome profiling comparison was made on the nodules of two soybean cultivars that differ in sensitivity to dark chilling [
46]. Microarray analysis was performed on nodules at seven weeks and 11 weeks, points where nitrogenase activity was maximal and where it had decreased to 50% of maximal rates [
49]. The cultivars used in this study were Highveld Top, which is nominally chilling resistant and PAN 809, which is nominally chilling sensitive. While only 10% of the mRNAs that showed age-dependent changes in abundance could be annotated, 220 showed a common developmental pattern in both soybean cultivars with 180 genes up-regulated and 40 genes down-regulated in the nodules at the early senescent stage [
49]. About 17% of up-regulated gene sequences belonged to transcription factor families. The dataset of senescence up-regulated genes also included those encoding enzymes involved in metabolism, such as cell wall modification and in general metabolic regulation, as well as stress responses [
49]. Seven of the identified gene sequences were involved in protein degradation and these were up-regulated in the nodules at the early senescent stage. These included a cysteine protease, two peptidases and four putative trypsin (serine protease) inhibitors. The cysteine protease (Gma.8481.1.S1_at) that was up-regulated in the nodules at the early senescent stage belongs to the group of vacuolar processing enzymes (VPEs). However, neither papain-like cysteine proteases nor cystatins were changed in expression in the soybean nodules of either cultivar at the early senescent stage.
The soybean genome data base [
50,
51,
52,
53,
54,
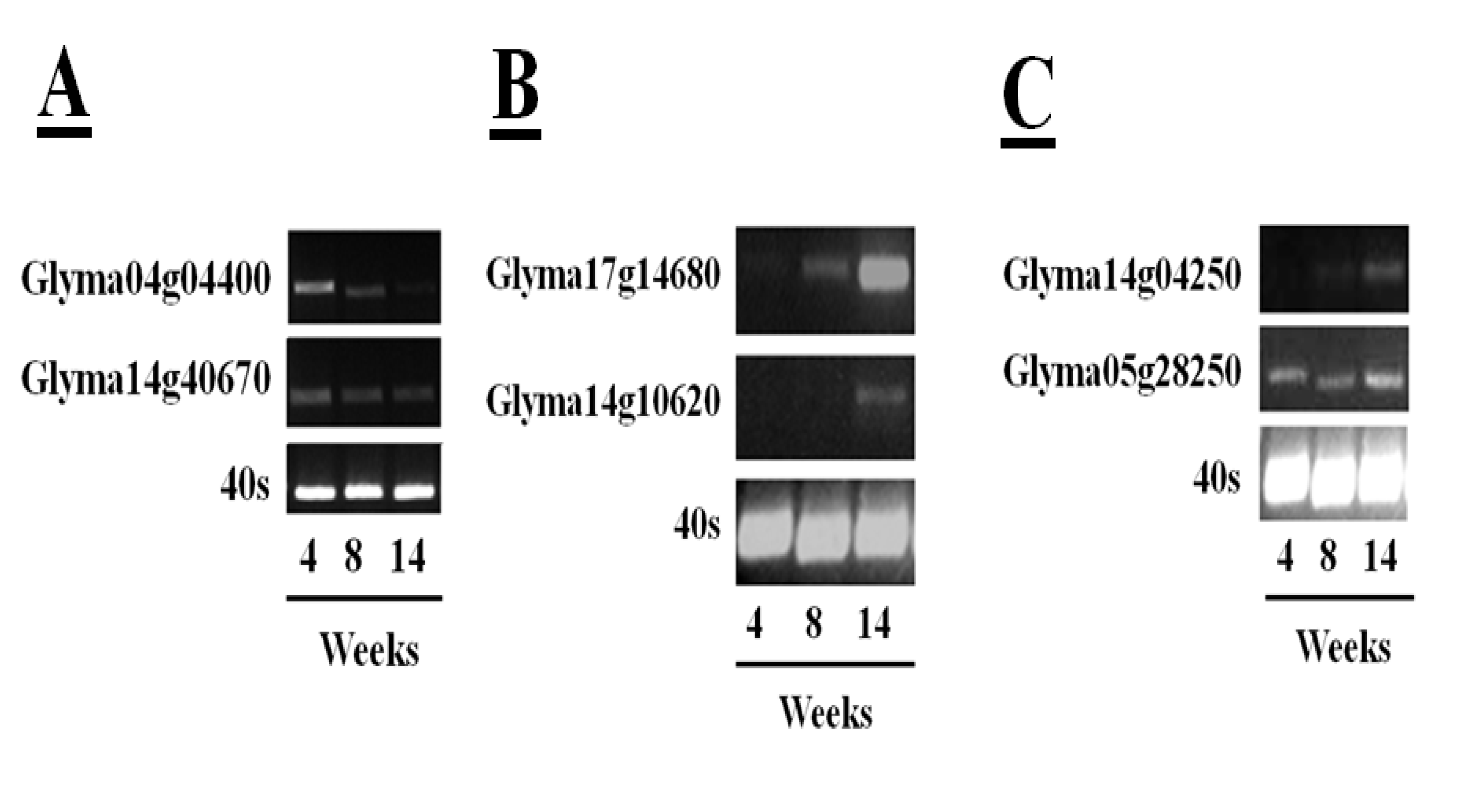
55] has facilitated rapid identification of soybean cysteine proteases and their inhibitors. The sequences in the databases include 52 putative papain-like cysteine protease gene sequences, eight legumain-like cysteine proteases and 18 putative gene sequences with homology to the rice cystatin-I. This information has allowed a more in-depth analysis of the changes in transcripts encoding the different proteins of the cysteine protease–cystatin system in soybean crown nodules during development. RNA sequencing (RNA-Seq) of crown nodules (authors own unpublished data) revealed that papain-like and legumain-like proteases (VPEs) as well as cystatins are transcribed in crown nodule. These genes were amplified and sequenced so that identity could be confirmed by alignment with sequences from other species using database comparisons using Blast N and ClustalW. Semi-quantitative real-time PCR analysis was performed on crown nodules harvested at different stages of development (four, eight, and 14 weeks) to detect amplified products that had 100% homology to accessions used as templates. Only two of the sequences that encode putative papain-like cysteine proteases produced visible amplification products after 26 PCR cycles (
Figure 6A,
Table 1), the other papain-like cysteine proteases tested requiring up to 40 cycles for production of a visible amplification product. These data suggest that the two papain-like cysteine proteases (Glyma04g04400 and Glyma14g40670) detected after 26 PCR cycles are the most abundant papain-like cysteine proteases in nodules. However, the abundance of these transcripts was not increased during senescence. A much greater amount of amplification products was found for the sequences encoding legumain-like cysteine proteases (Glyma17g14680 and Glyma14g10620). In contrast to young nodules, which yielded little or no detectable amplification, high amplification of these cDNAs was observed when 26 PCR cycles were applied to extracts from the 14 week-old nodules (
Figure 6B), suggesting that these transcripts were greatly increased in the senescent nodules. This result confirms a previous report showing increased expression of the legumain-like cysteine protease GmCysP1 in senescent nodules [
12]. A database search revealed that GMCysP1 is identical to Glyma17g14680. The expression of legumain-like cysteine proteases is likely to be required in order to release and activate the inactive cysteine proteases that have been stored in the vacuoles and other compartments of the nodules. Cystatin are also expressed during nodule development. In this case amplification products of only two cystatins, Glyma14g04250 and Glyma05g28250, were detectable after 30 PCR cycles (
Figure 6C), with some increase in the levels of Glyma14g04250 transcripts in senescent nodules.
Figure 6.
Transcription profiles of five papain-like cysteine proteases in soybean tissues of plants (cultivar Prima 2000) of different age (4, 8, and 14 weeks) determined by semi-quantitative RT-PCR. Amplification of the 40S gene sequence was used as a control. Semi-quantitative real-time polymerase chain reactions (semi-qRTPCR) were performed using 200 ng cDNA. For the reaction, 1 µL of the synthesized cDNA was used in a PCR reaction containing 2 µL of 10× PCR buffer, 1.6 µL of 25 mM MgCl2, 2 µL of 2 mM deoxyribonucleotide triphosphates (dNTPs), 0.4 µL of primers (10 µM), 0.2 µL of five units Taq polymerase. DNA amplification was done in a thermal cycler (Bio-Rad, USA) with denaturing DNA at 94 °C for 5 min followed by 25 to 46 cycles depending on the amplification product consisting of 94 °C for 30 s, 55 °C for 30 s for DNA annealing and 72 °C for 1 min for DNA extension. Final DNA elongation at 72 °C for 10 min. PCR samples were run on a 2% (w/v) agarose gel and DNA was stained with GelRed™ (Biotum, USA) added to 6X Loading Dye (Fermentas, Canada).
Figure 6.
Transcription profiles of five papain-like cysteine proteases in soybean tissues of plants (cultivar Prima 2000) of different age (4, 8, and 14 weeks) determined by semi-quantitative RT-PCR. Amplification of the 40S gene sequence was used as a control. Semi-quantitative real-time polymerase chain reactions (semi-qRTPCR) were performed using 200 ng cDNA. For the reaction, 1 µL of the synthesized cDNA was used in a PCR reaction containing 2 µL of 10× PCR buffer, 1.6 µL of 25 mM MgCl2, 2 µL of 2 mM deoxyribonucleotide triphosphates (dNTPs), 0.4 µL of primers (10 µM), 0.2 µL of five units Taq polymerase. DNA amplification was done in a thermal cycler (Bio-Rad, USA) with denaturing DNA at 94 °C for 5 min followed by 25 to 46 cycles depending on the amplification product consisting of 94 °C for 30 s, 55 °C for 30 s for DNA annealing and 72 °C for 1 min for DNA extension. Final DNA elongation at 72 °C for 10 min. PCR samples were run on a 2% (w/v) agarose gel and DNA was stained with GelRed™ (Biotum, USA) added to 6X Loading Dye (Fermentas, Canada).
![Agronomy 03 00550 g006]()
Table 1.
Forward and reverse primers used for amplification of different papain- and legumain-like cysteine proteases and cystatins with size of amplicons produced and cycles required to obtain an amplification product.
Table 1.
Forward and reverse primers used for amplification of different papain- and legumain-like cysteine proteases and cystatins with size of amplicons produced and cycles required to obtain an amplification product.
| Accession number | Forward Primer | Reverse Primer | Amplicon (bp) | Cycles |
|---|
Papain-like
Glyma04g04400 | GATCTTTAATGGCCACGATCCTCAT | CAGCACCTTGAAAGGGGTAATCCT | 678 | 26 |
| Glyma14g40670 | ATATGGAGCGTGTGACTCGG | GTAATATCCATTCTCTCCCCAGCTC | 430 | 26 |
Legumain-like
Glyma17g14680 | CTACGGAAACTACAGGCATC | GTTCTCCGTCGTCACATTAT | 217 | 26 |
Glyma14g10620
Cystatins | GGTCGTGGATGTTGCTGAGG | ATCTGCTTGATGCCTGTAGTTTCC | 191 | 26 |
| Glyma14g04250 | CACCGAAAGAGGATTAACAG | GGAGTTTGTGAGGGTGATTA | 180 | 30 |
| Glyma05g28250 | GGATTAAAGCATACTAAACCA | GAATATTCGAATCCGTTGT | 161 | 30 |
5. Characterization of the Soybean Nodule Cysteine Protease Proteome
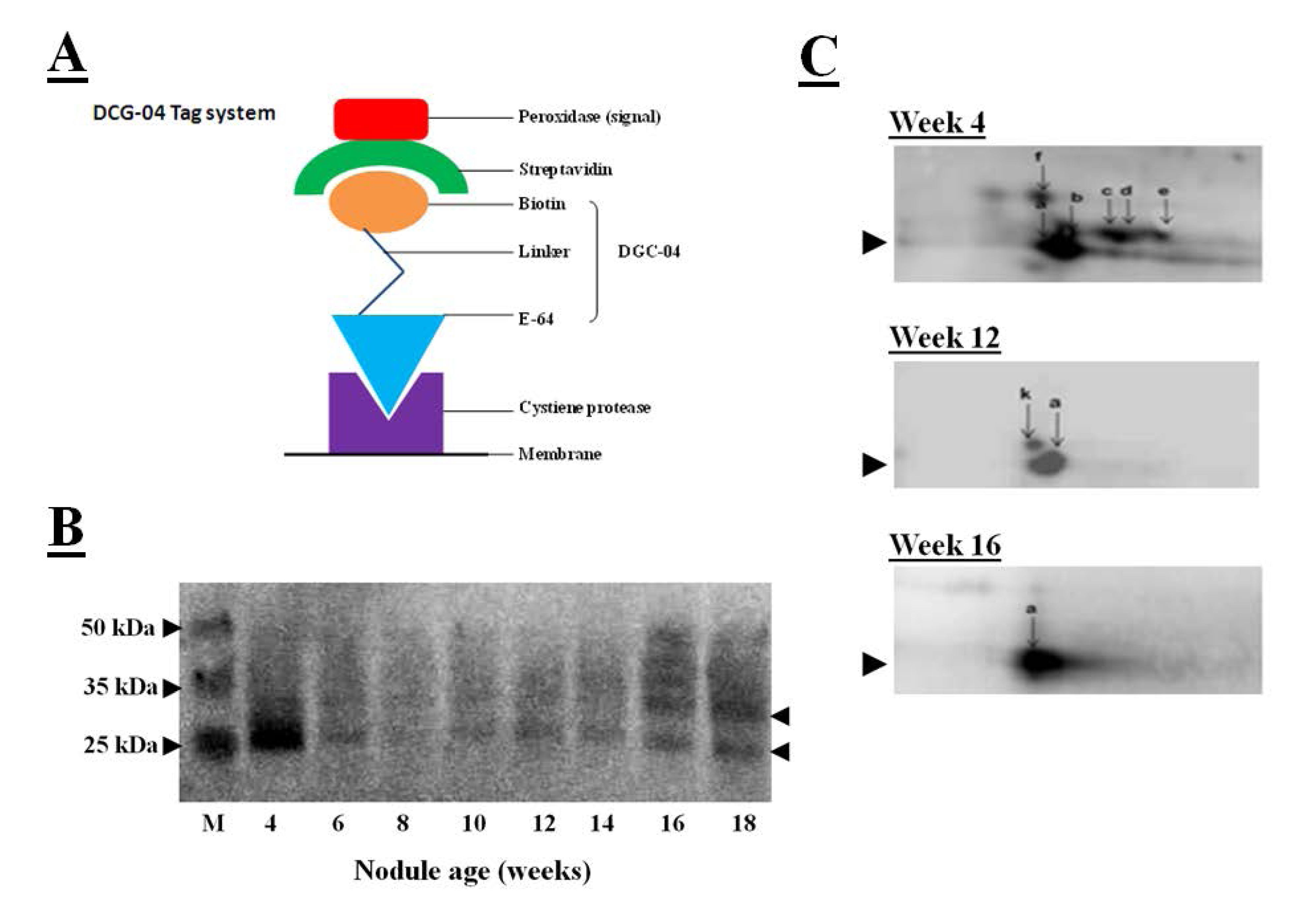
When combing with measurements of the hydrolysis of protease substrates, the transcriptomics data provide a wealth of data allowing the identification of the protease genes and their substrates. However, protease activity often not depends on translation of transcripts into proteins but also post-translational modifications of proteins. The DCG-04 tagging technique, used together with visualization of tagged proteases by a streptavidin-based detection system, can assist the accurate identification of proteases and their substrates [
56]. DCG-04 is an analog of the papain-like cysteine protease inhibitor E-64 that binds to these enzymes in an irreversible manner. DCG-04 carries a biotin residue allowing the tagging of substrate cysteine proteases by detection of the biotin carrying tagged proteins with peroxidase-labeled streptavidin, as illustrated in
Figure 7A. Like E64, DCG-04 reacts with active cysteine proteases in a mechanism-based manner. Enrichment of the papain-like cysteine protease proteome using this tagging technique, first by incubation with DCG-04, followed by the separation of tagged proteases by SDS-polyacrylamide gel electrophoresis and blotting the DCG-04-tagged proteins onto membranes and identification using a chemiluminescene substrate, revealed DCG-04 tagged protein bands with a molecular mass of between 25 and 35 kDa that were expressed in crown nodule during development, as illustrated in
Figure 7B. Detection in a one-dimensional SDS gel system revealed a strongly tagged band in extracts from four-week-old nodules. This band could not be related to an increase in total protease activity (
Figure 3A) or to an increase in cathepsin-L like activity (
Figure 4A). However, tagged proteins with weaker detection signals were more intense in the senescent 16 and 18 week-old nodules.
Figure 7.
(
A) Streptavidin-based detection system for cysteine proteases where peroxidase-labeled streptavidin binds to biotin linked with DGC-04 which is an E-64 analogue. DGC-04 binds to the active site of the cysteine proteases allowing detection of DCG-04-bound cysteine proteases via peroxidase activity measurement; (
B) Detection of DCG-04-labeled cysteine proteases produced in 4 to 18 weeks old soybean crown nodules (cultivar Prima 2000). Arrows on the right indicate position of detected cysteine protease bands. M represents different size protein markers. For labelling and detection of DCG-04 bound cysteine proteases with a streptavidin-based detection system the method described by Martinez
et al. [
57] was applied with five plant samples pooled; (
C) Two-dimensional PAGE for detection of DCG-04-labeled cysteine proteases produced in 4 to 16 weeks old crown nodules. For first dimension, acetone precipitated nodule proteins (40 µg) were first added to 130 µL of the sample rehydration solution (8 M urea, 2 M thiourea, 2% CHAPS as a detergent, 50 mM DTT as a reducing agent, 0.02% ampholyte solution, and 5% bromophenol blue as a dye) in order to denature and solubilise the sample proteins for isoelectric focusing using ZOOM IPG Runner system according to the supplier’s instructions (Life Technologies, UK). Second dimension was performed as SDS-PAGE according to Laemmli [
43] using a 15% polyacrylamide gel and Bio-Rad mini-protean III apparatus Bio-Rad, UK). Protein spots were blotted onto a nitrocellulose membrane and the membrane was developed using the Thermoscientific enhanced chemiluminescent kit according to manufacturer’s instructions (Thermo Fisher Scientific, USA) and visualized on a Syngene GeneSnap imaging system (Syngene, UK).
Figure 7.
(
A) Streptavidin-based detection system for cysteine proteases where peroxidase-labeled streptavidin binds to biotin linked with DGC-04 which is an E-64 analogue. DGC-04 binds to the active site of the cysteine proteases allowing detection of DCG-04-bound cysteine proteases via peroxidase activity measurement; (
B) Detection of DCG-04-labeled cysteine proteases produced in 4 to 18 weeks old soybean crown nodules (cultivar Prima 2000). Arrows on the right indicate position of detected cysteine protease bands. M represents different size protein markers. For labelling and detection of DCG-04 bound cysteine proteases with a streptavidin-based detection system the method described by Martinez
et al. [
57] was applied with five plant samples pooled; (
C) Two-dimensional PAGE for detection of DCG-04-labeled cysteine proteases produced in 4 to 16 weeks old crown nodules. For first dimension, acetone precipitated nodule proteins (40 µg) were first added to 130 µL of the sample rehydration solution (8 M urea, 2 M thiourea, 2% CHAPS as a detergent, 50 mM DTT as a reducing agent, 0.02% ampholyte solution, and 5% bromophenol blue as a dye) in order to denature and solubilise the sample proteins for isoelectric focusing using ZOOM IPG Runner system according to the supplier’s instructions (Life Technologies, UK). Second dimension was performed as SDS-PAGE according to Laemmli [
43] using a 15% polyacrylamide gel and Bio-Rad mini-protean III apparatus Bio-Rad, UK). Protein spots were blotted onto a nitrocellulose membrane and the membrane was developed using the Thermoscientific enhanced chemiluminescent kit according to manufacturer’s instructions (Thermo Fisher Scientific, USA) and visualized on a Syngene GeneSnap imaging system (Syngene, UK).
![Agronomy 03 00550 g007]()
Two-dimensional polyacrylamide gel electrophoresis was also used to characterize the papain-like DCG-04 tagged cysteine proteases further. In these studies, tagged cysteine proteases were blotted onto membranes and detected by the streptavidin-peroxidase system. In this system, the major band that had been detected in young nodules using one-dimensional polyacrylamide gel electrophoresis, separated into several spots that represent different papain-like cysteine proteases with a molecular mass of about 30 kDa (
Figure 7C). The number of detectable spots was greatest in the young nodules and progressively decreased as the nodules senesced such that only one major spot was detectable in the 16 week-old nodules. This analysis indicates that a number of papain-like cysteine proteases are present at the early stages of nodule development. Further analysis is required to determine the identities of these proteases and their substrates. To date, the enrichment of cystatins by using the streptavidin-tagged papain has not been successful.
7. Conclusions
There is accumulating evidence that proteases, including cysteine proteases, are produced during legume nodule development. The data presented here have been used to illustrate that cysteine proteases are present at all stages of nodule development but that the composition of expressed forms is different in senescent nodules from that in young nodules. The expression and hence function of certain cysteine proteases is specific to nodule senescence allowing protein degradation and loss of nitrogen fixation. The data presented here indicate that legumain-like cysteine proteases, which are required to release and active papain-like cysteine proteases, are the major cysteine proteases that are expressed in senescent nodules, as illustrated in
Figure 8, which provides a schematic overview of expression changes for the cysteine protease–cystatin system in during soybean nodule development.
Several papain-like cysteine proteases are expressed in young nitrogen fixing nodules but this expression does not appear to greatly alter the overall cysteine protease activity of the nodules. There is no evidence to date concerning cystatin involvement in soybean nodule development. We have, as yet, been unable to find any significant increases in the transcription of cystatins during nodule development. Cystatins fulfill important roles in biotic stress protection [
11]. It is possible therefore to speculate that they may function in nodule development to prevent the papain-like cysteine protease-mediated proteolysis in nodules exposed to stress that would lead to stress-induced senescence. The finding that the ectopic over-expressing the rice cystatin-I in transgenic tobacco plants allowed a better recovery of photosynthesis after exposure to cold stress supports this view [
62]. There is also evidence that some phytocystatins contain a carboxy-terminal extension with an amino acid motif (SNSL) similar to that involved in the inhibition of legumain-like proteins by human cystatins [
63]. We have already identified phytocystatins in soybean nodules and we are currently testing the hypothesis that these cystatins might act as legumain inhibitors regulating legumain activity during nodule development. The above discussion has illustrated the fact that very little is currently known about cystatin function in nodules and that further research is vital to determine the role of the complete cysteine protease–cystatin system in nodule development under optimal and stress conditions.
Figure 8.
Schematic diagram of different types of cysteine proteases (CP) and cystatins expressed in soybean nodules at different development stages.
Figure 8.
Schematic diagram of different types of cysteine proteases (CP) and cystatins expressed in soybean nodules at different development stages.